Structural insights into the inactivation of CRISPR-Cas systems by diverse anti-CRISPR proteins
- PMID: 29554913
- PMCID: PMC5859409
- DOI: 10.1186/s12915-018-0504-9
Structural insights into the inactivation of CRISPR-Cas systems by diverse anti-CRISPR proteins
Abstract
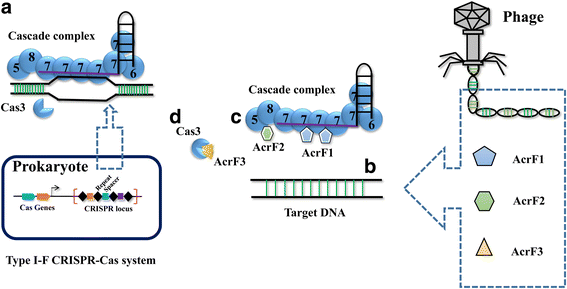
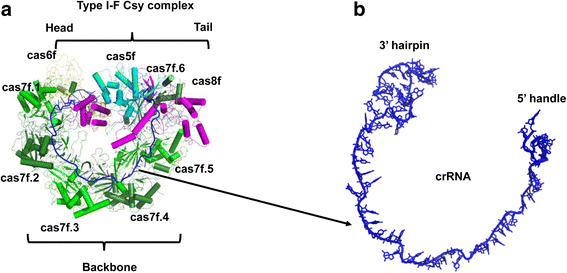
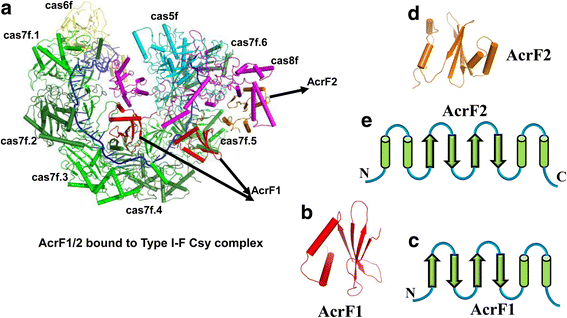
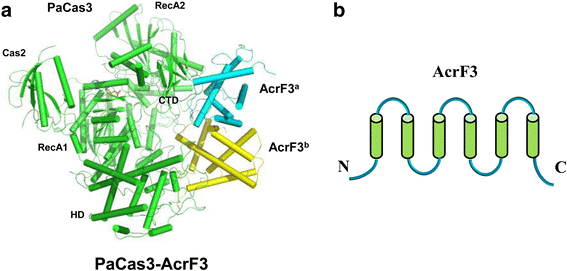
A molecular arms race is progressively being unveiled between prokaryotes and viruses. Prokaryotes utilize CRISPR-mediated adaptive immune systems to kill the invading phages and mobile genetic elements, and in turn, the viruses evolve diverse anti-CRISPR proteins to fight back. The structures of several anti-CRISPR proteins have now been reported, and here we discuss their structural features, with a particular emphasis on topology, to discover their similarities and differences. We summarize the CRISPR-Cas inhibition mechanisms of these anti-CRISPR proteins in their structural context. Considering anti-CRISPRs in this way will provide important clues for studying their origin and evolution.
Keywords: Adaptive immune; Anti-CRISPR; CRISPR-Cas system; Structure; Viral infection.
Conflict of interest statement
Competing interest
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
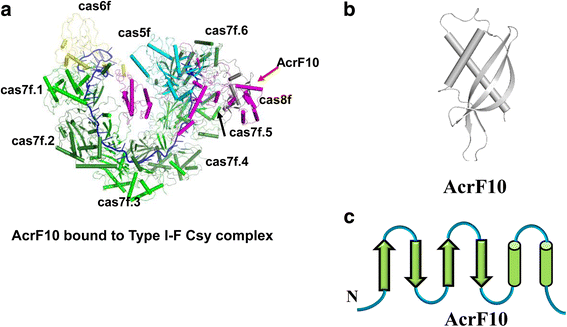
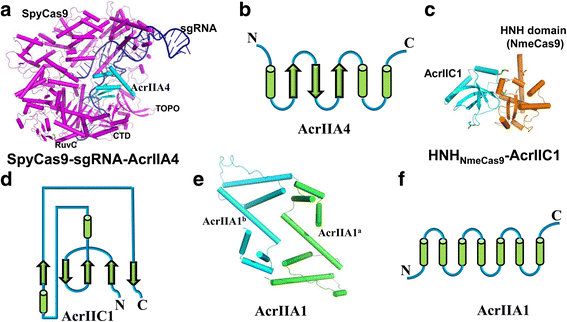
Similar articles
-
CRISPR-Cas adaptation: insights into the mechanism of action.Nat Rev Microbiol. 2016 Feb;14(2):67-76. doi: 10.1038/nrmicro.2015.14. Epub 2016 Jan 11. Nat Rev Microbiol. 2016. PMID: 26751509 Review.
-
Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins.Nature. 2015 Oct 1;526(7571):136-9. doi: 10.1038/nature15254. Epub 2015 Sep 23. Nature. 2015. PMID: 26416740 Free PMC article.
-
Adaptation in CRISPR-Cas Systems.Mol Cell. 2016 Mar 17;61(6):797-808. doi: 10.1016/j.molcel.2016.01.030. Epub 2016 Mar 3. Mol Cell. 2016. PMID: 26949040 Review.
-
[Why so rare if so essentiel: the determinants of the sparse distribution of CRISPR-Cas systems in bacterial genomes].Biol Aujourdhui. 2017;211(4):255-264. doi: 10.1051/jbio/2018005. Epub 2018 Jun 29. Biol Aujourdhui. 2017. PMID: 29956652 Review. French.
-
Inhibition of CRISPR-Cas systems by mobile genetic elements.Curr Opin Microbiol. 2017 Jun;37:120-127. doi: 10.1016/j.mib.2017.06.003. Epub 2017 Jun 29. Curr Opin Microbiol. 2017. PMID: 28668720 Free PMC article. Review.
Cited by
-
A high-resolution (1.2 Å) crystal structure of the anti-CRISPR protein AcrIF9.FEBS Open Bio. 2020 Dec;10(12):2532-2540. doi: 10.1002/2211-5463.12986. Epub 2020 Nov 5. FEBS Open Bio. 2020. PMID: 32990416 Free PMC article.
-
Anti-CRISPRs: The natural inhibitors for CRISPR-Cas systems.Animal Model Exp Med. 2019 May 29;2(2):69-75. doi: 10.1002/ame2.12069. eCollection 2019 Jun. Animal Model Exp Med. 2019. PMID: 31392299 Free PMC article. Review.
-
In silico analysis reveals the co-existence of CRISPR-Cas type I-F1 and type I-F2 systems and its association with restricted phage invasion in Acinetobacter baumannii.Front Microbiol. 2022 Aug 17;13:909886. doi: 10.3389/fmicb.2022.909886. eCollection 2022. Front Microbiol. 2022. PMID: 36060733 Free PMC article.
-
Molecular basis of anti-CRISPR operon repression by Aca10.Nucleic Acids Res. 2022 Aug 26;50(15):8919-8928. doi: 10.1093/nar/gkac656. Nucleic Acids Res. 2022. PMID: 35920325 Free PMC article.
-
Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.Nucleic Acids Res. 2020 Sep 25;48(17):9959-9968. doi: 10.1093/nar/gkaa690. Nucleic Acids Res. 2020. PMID: 32810226 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

