Induction of apoptosis in imatinib sensitive and resistant chronic myeloid leukemia cells by efficient disruption of bcr-abl oncogene with zinc finger nucleases
- PMID: 29554925
- PMCID: PMC5859405
- DOI: 10.1186/s13046-018-0732-4
Induction of apoptosis in imatinib sensitive and resistant chronic myeloid leukemia cells by efficient disruption of bcr-abl oncogene with zinc finger nucleases
Abstract
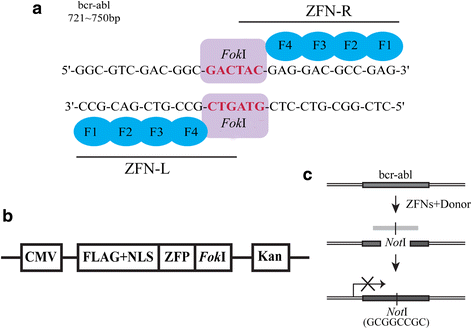
Background: The bcr-abl fusion gene is the pathological origin of chronic myeloid leukemia (CML) and plays a critical role in the resistance of imatinib. Thus, bcr-abl disruption-based novel therapeutic strategy may warrant exploration. In our study, we were surprised to find that the characteristics of bcr-abl sequences met the design requirements of zinc finger nucleases (ZFNs).
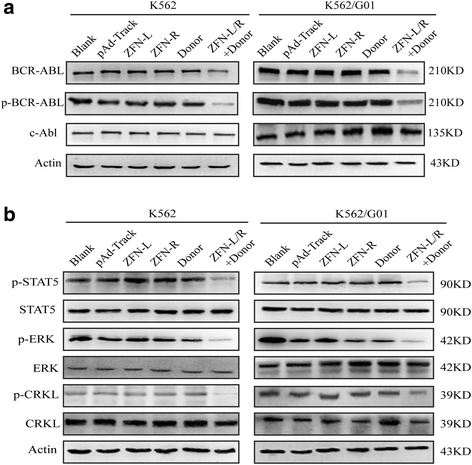
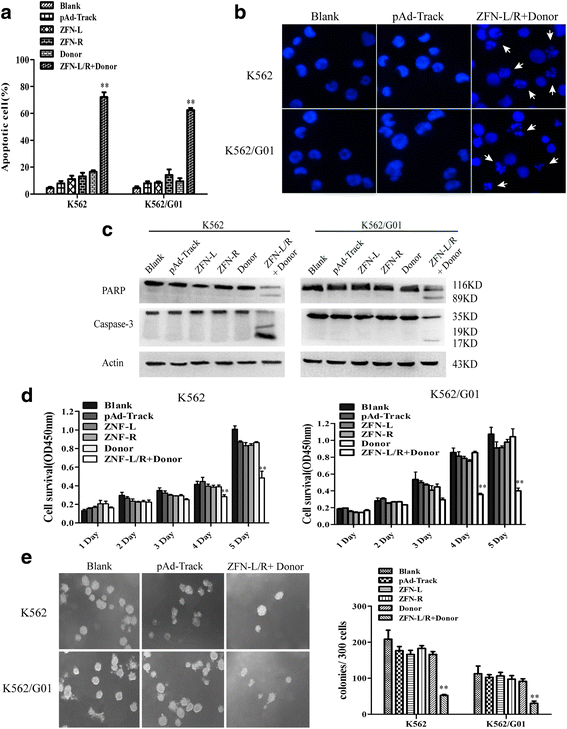
Methods: We constructed the ZFNs targeting bcr-abl with high specificity through simple modular assembly approach. Western blotting was conducted to detect the expression of BCR-ABL and phosphorylation of its downstream STAT5, ERK and CRKL in CML cells. CCK8 assay, colony-forming assay and flow cytometry (FCM) were used to evaluate the effect of the ZFNs on the viablity and apoptosis of CML cells and CML CD34+ cells. Moreover, mice model was used to determine the ability of ZFNs in disrupting the leukemogenesis of bcr-abl in vivo.
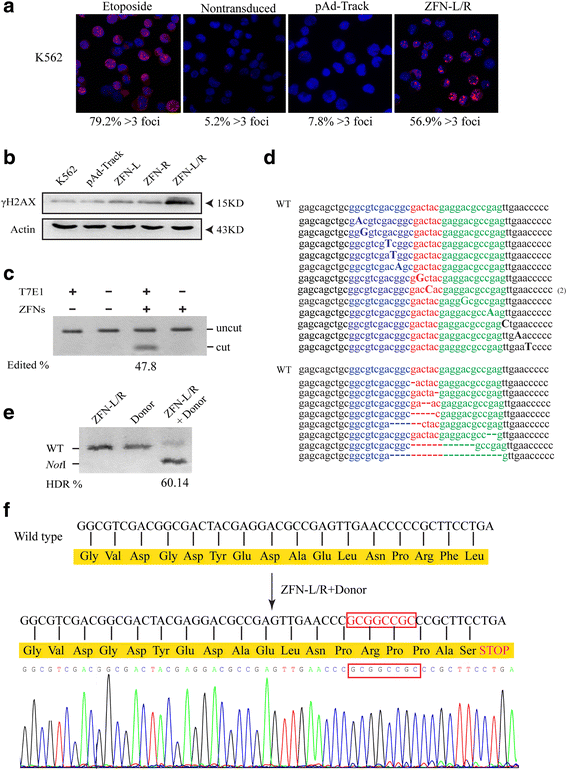
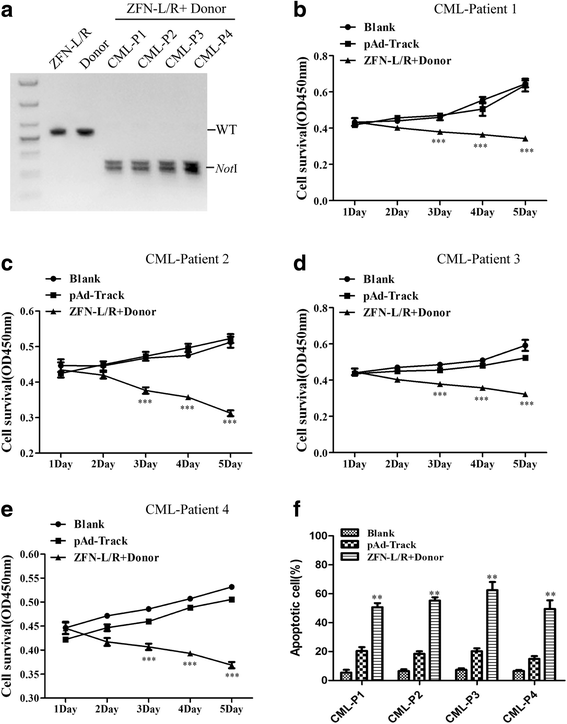
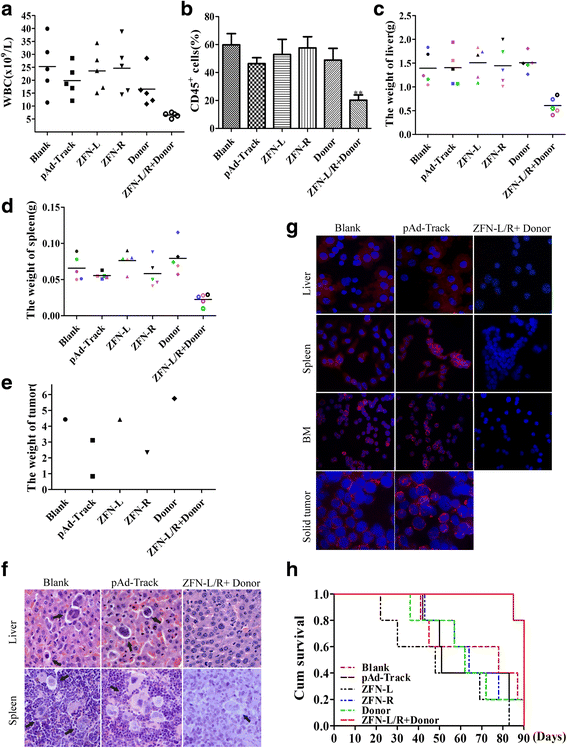
Results: The ZFNs skillfully mediated 8-base NotI enzyme cutting site addition in bcr-abl gene of imatinib sensitive and resistant CML cells by homology-directed repair (HDR), which led to a stop codon and terminated the translation of BCR-ABL protein. As expected, the disruption of bcr-abl gene induced cell apoptosis and inhibited cell proliferation. Notably, we obtained similar result in CD34+ cells from CML patients. Moreover, the ZFNs significantly reduced the oncogenicity of CML cells in mice.
Conclusion: These results reveal that the bcr-abl gene disruption based on ZFNs may provide a treatment choice for imatinib resistant or intolerant CML patients.
Keywords: Bcr-abl; Chronic myeloid leukemia; Homology-directed repair; Oncogenicity; Zinc finger nucleases.
Conflict of interest statement
Ethics approval and consent to participate
All samples were collected with informed consent and all the experiment were approved by the ethical committee. All animal experiments were in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) and were conducted with the approval of the Biomedical Ethics Committee of Chongqing Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
MeSH terms
Substances
Grants and funding
- 81572060/National Natural Science Foundation of China
- 81500129/National Natural Science Foundation of China
- 201415/Research and Cultivation Fund Project of Chongqing Medical University
- KJ1500215/Scientific and Technological Research Program of Chongqing Municipal Education Commission
- No.CYS15120/Chongqing Postgraduate Research and innovation project
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

