Species classifier choice is a key consideration when analysing low-complexity food microbiome data
- PMID: 29554948
- PMCID: PMC5859664
- DOI: 10.1186/s40168-018-0437-0
Species classifier choice is a key consideration when analysing low-complexity food microbiome data
Abstract
Background: The use of shotgun metagenomics to analyse low-complexity microbial communities in foods has the potential to be of considerable fundamental and applied value. However, there is currently no consensus with respect to choice of species classification tool, platform, or sequencing depth. Here, we benchmarked the performances of three high-throughput short-read sequencing platforms, the Illumina MiSeq, NextSeq 500, and Ion Proton, for shotgun metagenomics of food microbiota. Briefly, we sequenced six kefir DNA samples and a mock community DNA sample, the latter constructed by evenly mixing genomic DNA from 13 food-related bacterial species. A variety of bioinformatic tools were used to analyse the data generated, and the effects of sequencing depth on these analyses were tested by randomly subsampling reads.
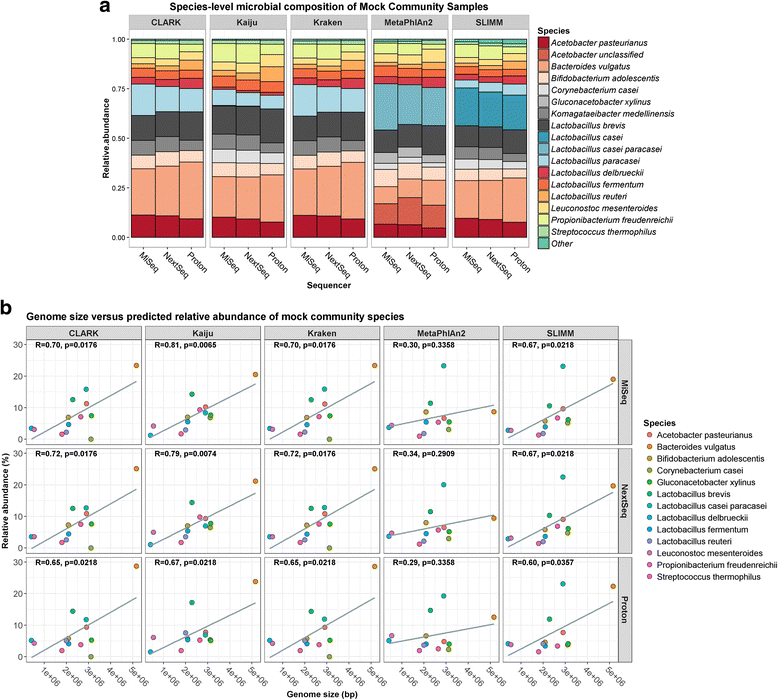
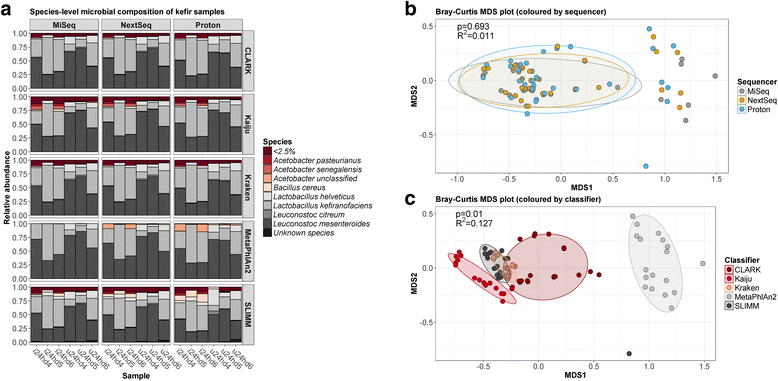
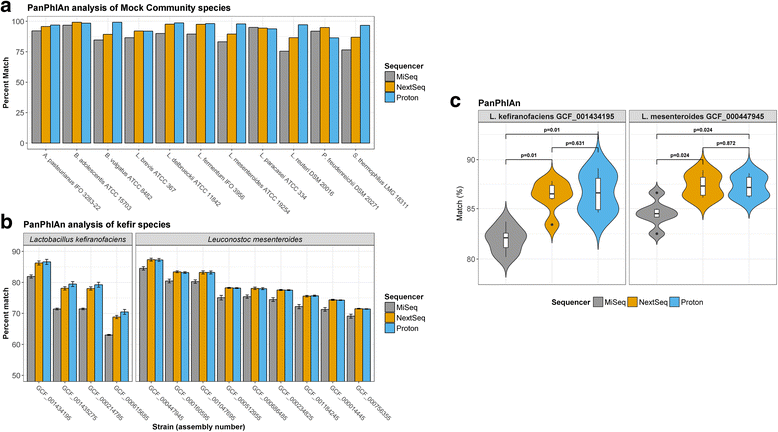
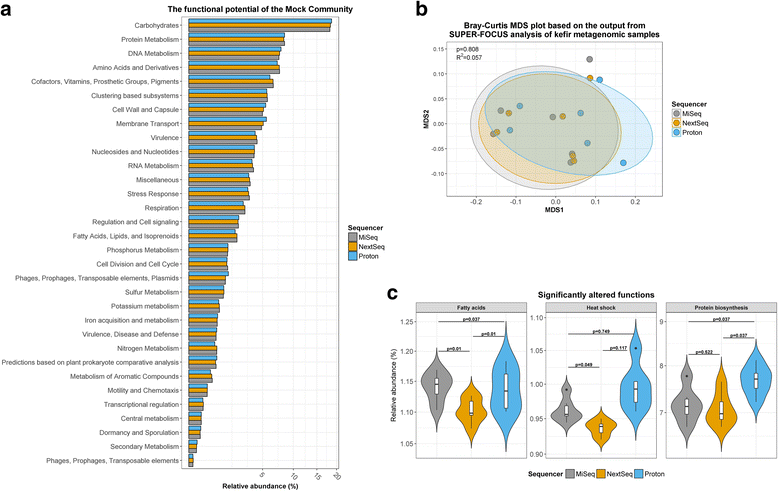
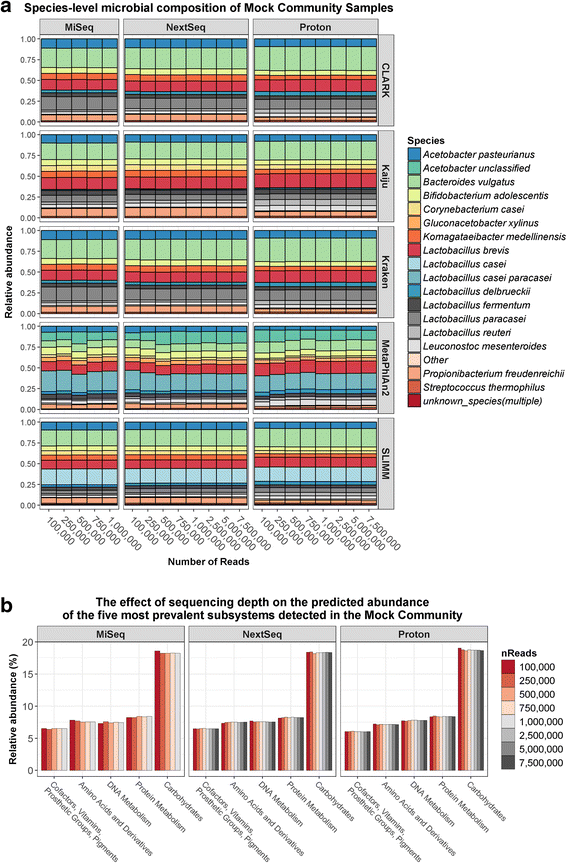
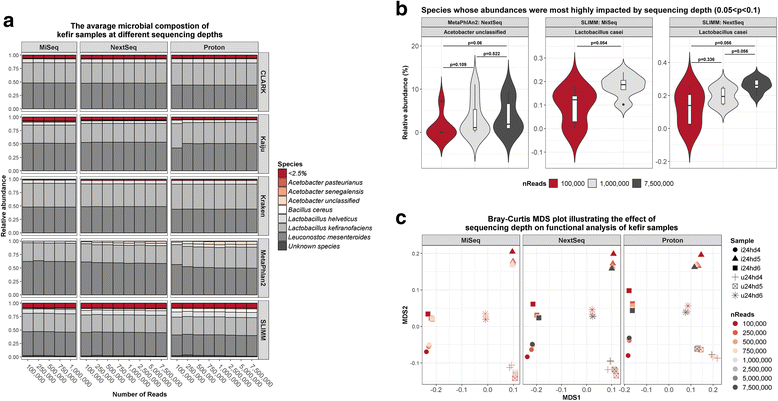
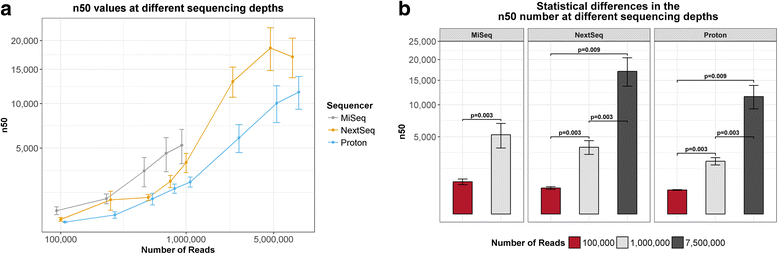
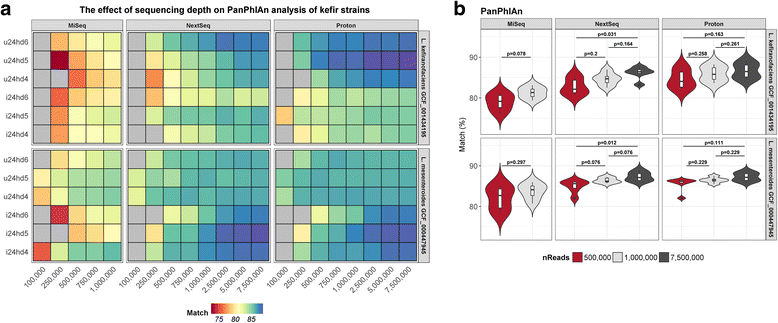
Results: Compositional analysis results were consistent between the platforms at divergent sequencing depths. However, we observed pronounced differences in the predictions from species classification tools. Indeed, PERMANOVA indicated that there was no significant differences between the compositional results generated by the different sequencers (p = 0.693, R2 = 0.011), but there was a significant difference between the results predicted by the species classifiers (p = 0.01, R2 = 0.127). The relative abundances predicted by the classifiers, apart from MetaPhlAn2, were apparently biased by reference genome sizes. Additionally, we observed varying false-positive rates among the classifiers. MetaPhlAn2 had the lowest false-positive rate, whereas SLIMM had the greatest false-positive rate. Strain-level analysis results were also similar across platforms. Each platform correctly identified the strains present in the mock community, but accuracy was improved slightly with greater sequencing depth. Notably, PanPhlAn detected the dominant strains in each kefir sample above 500,000 reads per sample. Again, the outputs from functional profiling analysis using SUPER-FOCUS were generally accordant between the platforms at different sequencing depths. Finally, and expectedly, metagenome assembly completeness was significantly lower on the MiSeq than either on the NextSeq (p = 0.03) or the Proton (p = 0.011), and it improved with increased sequencing depth.
Conclusions: Our results demonstrate a remarkable similarity in the results generated by the three sequencing platforms at different sequencing depths, and, in fact, the choice of bioinformatics methodology had a more evident impact on results than the choice of sequencer did.
Keywords: Low-complexity microbiome; Sequencing platform comparison; Shotgun metagenomics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

