Esx Paralogs Are Functionally Equivalent to ESX-1 Proteins but Are Dispensable for Virulence in Mycobacterium marinum
- PMID: 29555701
- PMCID: PMC5952400
- DOI: 10.1128/JB.00726-17
Esx Paralogs Are Functionally Equivalent to ESX-1 Proteins but Are Dispensable for Virulence in Mycobacterium marinum
Abstract
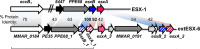
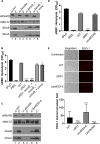
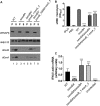
Mycobacterium marinum is a nontuberculous pathogen of poikilothermic fish and an opportunistic human pathogen. Like tuberculous mycobacteria, the M. marinum M strain requires the ESX-1 (ESAT-6 system 1) secretion system for virulence in host cells. EsxB and EsxA, two major virulence factors exported by the ESX-1 system, are encoded by the esxBA genes within the ESX-1 locus. Deletion of the esxBA genes abrogates ESX-1 export and attenuates M. marinum in ex vivo and in vivo models of infection. Interestingly, there are several duplications of the esxB and esxA genes (esxB_1, esxB_2, esxA_1, esxA_2, and esxA_3) in the M. marinum M genome located outside the ESX-1 locus. We sought to understand if this region, known as ESX-6, contributes to ESX-1-mediated virulence. We found that deletion of the esxB_1 gene alone or the entire ESX-6 locus did not impact ESX-1 export or function, supporting the idea that the esxBA genes present at the ESX-1 locus are the primary contributors to ESX-1-mediated virulence. Nevertheless, overexpression of the esxB_1 locus complemented ESX-1 function in the ΔesxBA strain, signifying that the two loci are functionally equivalent. Our findings raise questions about why duplicate versions of the esxBA genes are maintained in the M. marinum M genome and how these proteins, which are functionally equivalent to virulence factors, contribute to mycobacterial biology.IMPORTANCEMycobacterium tuberculosis is the causative agent of the human disease tuberculosis (TB). There are 10.4 million cases and 1.7 million TB-associated deaths annually, making TB a leading cause of death globally. Nontuberculous mycobacteria (NTM) cause chronic human infections that are acquired from the environment. Despite differences in disease etiology, both tuberculous and NTM pathogens use the ESX-1 secretion system to cause disease. The nontubercular mycobacterial species, Mycobacterium marinum, has additional copies of specific ESX-1 genes. Our findings demonstrate that the duplicated genes do not contribute to virulence but can substitute for virulence factors in M. marinum These findings suggest that the duplicated genes may play a specific role in NTM biology.
Keywords: CFP-10; ESX-1; ESX-6; duplication; gene duplication; mycobacteria; type VII secretion.
Copyright © 2018 American Society for Microbiology.
Figures

Similar articles
-
The ESX-1 Virulence Factors Downregulate miR-147-3p in Mycobacterium marinum-Infected Macrophages.Infect Immun. 2020 May 20;88(6):e00088-20. doi: 10.1128/IAI.00088-20. Print 2020 May 20. Infect Immun. 2020. PMID: 32253249 Free PMC article.
-
A novel ESX-1 locus reveals that surface-associated ESX-1 substrates mediate virulence in Mycobacterium marinum.J Bacteriol. 2014 May;196(10):1877-88. doi: 10.1128/JB.01502-14. Epub 2014 Mar 7. J Bacteriol. 2014. PMID: 24610712 Free PMC article.
-
Conserved ESX-1 Substrates EspE and EspF Are Virulence Factors That Regulate Gene Expression.Infect Immun. 2020 Nov 16;88(12):e00289-20. doi: 10.1128/IAI.00289-20. Print 2020 Nov 16. Infect Immun. 2020. PMID: 32900815 Free PMC article.
-
Modeling Tubercular ESX-1 Secretion Using Mycobacterium marinum.Microbiol Mol Biol Rev. 2020 Sep 2;84(4):e00082-19. doi: 10.1128/MMBR.00082-19. Print 2020 Nov 18. Microbiol Mol Biol Rev. 2020. PMID: 32878966 Free PMC article. Review.
-
Mycobacterium marinum as a model for understanding principles of mycobacterial pathogenesis.J Bacteriol. 2025 May 22;207(5):e0004725. doi: 10.1128/jb.00047-25. Epub 2025 Apr 30. J Bacteriol. 2025. PMID: 40304497 Free PMC article. Review.
Cited by
-
An ancestral mycobacterial effector promotes dissemination of infection.Cell. 2022 Nov 23;185(24):4507-4525.e18. doi: 10.1016/j.cell.2022.10.019. Epub 2022 Nov 9. Cell. 2022. PMID: 36356582 Free PMC article.
-
Infect and Inject: How Mycobacterium tuberculosis Exploits Its Major Virulence-Associated Type VII Secretion System, ESX-1.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.bai-0024-2019. doi: 10.1128/microbiolspec.BAI-0024-2019. Microbiol Spectr. 2019. PMID: 31172908 Free PMC article. Review.
-
The loss of the PDIM/PGL virulence lipids causes differential secretion of ESX-1 substrates in Mycobacterium marinum.mSphere. 2024 May 29;9(5):e0000524. doi: 10.1128/msphere.00005-24. Epub 2024 Apr 25. mSphere. 2024. PMID: 38661343 Free PMC article.
-
The 24th Annual Midwest Microbial Pathogenesis Meeting.J Bacteriol. 2018 Feb 26;200(11):e000950-18. doi: 10.1128/JB.00095-18. Online ahead of print. J Bacteriol. 2018. PMID: 29483166 Free PMC article.
-
A New ESX-1 Substrate in Mycobacterium marinum That Is Required for Hemolysis but Not Host Cell Lysis.J Bacteriol. 2019 Jun 21;201(14):e00760-18. doi: 10.1128/JB.00760-18. Print 2019 Jul 15. J Bacteriol. 2019. PMID: 30833360 Free PMC article.
References
-
- Feldman RA, Long MW, David HL. 1974. Mycobacterium marinum: a leisure-time pathogen. J Infect Dis 129:618–621. doi:10.1093/infdis/129.5.618. - DOI
-
- Aronson JD. 1926. Spontaneous tuberculosis in salt water fish. J Infect Dis 39:315–320. doi:10.1093/infdis/39.4.315. - DOI
-
- Koch R. 1952. Tuberculosis etiology. Dtsch Gesundheitsw 7:457–465. (Undetermined language.) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

