A pex1 missense mutation improves peroxisome function in a subset of Arabidopsis pex6 mutants without restoring PEX5 recycling
- PMID: 29555730
- PMCID: PMC5889664
- DOI: 10.1073/pnas.1721279115
A pex1 missense mutation improves peroxisome function in a subset of Arabidopsis pex6 mutants without restoring PEX5 recycling
Abstract
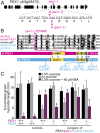
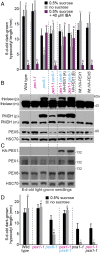
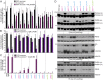
Peroxisomes are eukaryotic organelles critical for plant and human development because they house essential metabolic functions, such as fatty acid β-oxidation. The interacting ATPases PEX1 and PEX6 contribute to peroxisome function by recycling PEX5, a cytosolic receptor needed to import proteins targeted to the peroxisomal matrix. Arabidopsis pex6 mutants exhibit low PEX5 levels and defects in peroxisomal matrix protein import, oil body utilization, peroxisomal metabolism, and seedling growth. These defects are hypothesized to stem from impaired PEX5 retrotranslocation leading to PEX5 polyubiquitination and consequent degradation of PEX5 via the proteasome or of the entire organelle via autophagy. We recovered a pex1 missense mutation in a screen for second-site suppressors that restore growth to the pex6-1 mutant. Surprisingly, this pex1-1 mutation ameliorated the metabolic and physiological defects of pex6-1 without restoring PEX5 levels. Similarly, preventing autophagy by introducing an atg7-null allele partially rescued pex6-1 physiological defects without restoring PEX5 levels. atg7 synergistically improved matrix protein import in pex1-1 pex6-1, implying that pex1-1 improves peroxisome function in pex6-1 without impeding autophagy of peroxisomes (i.e., pexophagy). pex1-1 differentially improved peroxisome function in various pex6 alleles but worsened the physiological and molecular defects of a pex26 mutant, which is defective in the tether anchoring the PEX1-PEX6 hexamer to the peroxisome. Our results support the hypothesis that, beyond PEX5 recycling, PEX1 and PEX6 have additional functions in peroxisome homeostasis and perhaps in oil body utilization.
Keywords: AAA ATPase; oil bodies; peroxin; peroxisome; pexophagy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Disparate peroxisome-related defects in Arabidopsis pex6 and pex26 mutants link peroxisomal retrotranslocation and oil body utilization.Plant J. 2017 Oct;92(1):110-128. doi: 10.1111/tpj.13641. Epub 2017 Aug 22. Plant J. 2017. PMID: 28742939 Free PMC article.
-
The PEX1 ATPase Stabilizes PEX6 and Plays Essential Roles in Peroxisome Biology.Plant Physiol. 2017 Aug;174(4):2231-2247. doi: 10.1104/pp.17.00548. Epub 2017 Jun 9. Plant Physiol. 2017. PMID: 28600347 Free PMC article.
-
Peroxisomal ubiquitin-protein ligases peroxin2 and peroxin10 have distinct but synergistic roles in matrix protein import and peroxin5 retrotranslocation in Arabidopsis.Plant Physiol. 2014 Nov;166(3):1329-44. doi: 10.1104/pp.114.247148. Epub 2014 Sep 11. Plant Physiol. 2014. PMID: 25214533 Free PMC article.
-
Pexophagy is responsible for 65% of cases of peroxisome biogenesis disorders.Autophagy. 2017 May 4;13(5):991-994. doi: 10.1080/15548627.2017.1291480. Epub 2017 Feb 28. Autophagy. 2017. PMID: 28318378 Free PMC article. Review.
-
A Mechanistic Perspective on PEX1 and PEX6, Two AAA+ Proteins of the Peroxisomal Protein Import Machinery.Int J Mol Sci. 2019 Oct 23;20(21):5246. doi: 10.3390/ijms20215246. Int J Mol Sci. 2019. PMID: 31652724 Free PMC article. Review.
Cited by
-
Ubiquitin-conjugating activity by PEX4 is required for efficient protein transport to peroxisomes in Arabidopsis thaliana.J Biol Chem. 2022 Jun;298(6):102038. doi: 10.1016/j.jbc.2022.102038. Epub 2022 May 17. J Biol Chem. 2022. PMID: 35595097 Free PMC article.
-
Dynamics of Peroxisome Homeostasis and Its Role in Stress Response and Signaling in Plants.Front Plant Sci. 2019 Jun 4;10:705. doi: 10.3389/fpls.2019.00705. eCollection 2019. Front Plant Sci. 2019. PMID: 31214223 Free PMC article. Review.
-
The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane.Front Cell Dev Biol. 2022 Feb 18;10:838923. doi: 10.3389/fcell.2022.838923. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35300425 Free PMC article.
-
Insights into the Structure and Function of the Pex1/Pex6 AAA-ATPase in Peroxisome Homeostasis.Cells. 2022 Jun 29;11(13):2067. doi: 10.3390/cells11132067. Cells. 2022. PMID: 35805150 Free PMC article. Review.
-
A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes.Autophagy. 2019 Jun;15(6):941-959. doi: 10.1080/15548627.2019.1569915. Epub 2019 Feb 8. Autophagy. 2019. PMID: 30734619 Free PMC article.
References
-
- Bartel B, Burkhart SE, Fleming WA. Protein transport in and out of plant peroxisomes. In: Brocard C, Hartig A, editors. Molecular Machines Involved in Peroxisome Biogenesis and Maintenance. Springer; Vienna: 2014. pp. 325–345.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

