The role of PTRF/Cavin1 as a biomarker in both glioma and serum exosomes
- PMID: 29556340
- PMCID: PMC5858166
- DOI: 10.7150/thno.22952
The role of PTRF/Cavin1 as a biomarker in both glioma and serum exosomes
Abstract
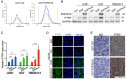
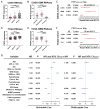
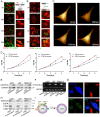
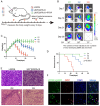
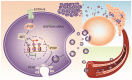
Exosomes play critical roles in intercellular communication in both nearby and distant cells in individuals and organs. Polymerase I and transcript release factor (PTRF), also known as Cavin1, has previously been described as a critical factor in caveola formation, and aberrant PTRF expression has been reported in various malignancies. However, the function of PTRF in tumor progression remains controversial, and its role in glioma is poorly understood. In this study, we report that PTRF is associated with malignancy grade and poor prognosis in glioma patients. Our previous study using two proteomics methods, tandem mass tag (TMT) and data-independent acquisition (DIA), showed that EGFRvIII overexpression increased PTRF expression at the protein level. In contrast, blocking PI3K and AKT using LY294002 and MK-2206, respectively, decreased PTRF expression, showing that PTRF is regulated in the EGFR/PI3K/AKT pathway. ChIP-PCR analysis showed that PTRF is transcriptionally regulated by the H3K4me3 and H3K27me3 modifications. Furthermore, PTRF overexpression increased exosome secretion and induced cell growth in vitro. More importantly, overexpressing PTRF induced the malignancy of nearby cells in vivo, suggesting that PTRF alters the microenvironment through intercellular communication via exosomes. Furthermore, analysis of clinical samples showed a positive correlation between tumor grade and PTRF expression in both tumor tissues and exosomes isolated from blood harvested from glioma patients, and PTRF expression in exosomes isolated from the sera of GBM patients was decreased after surgery. In conclusion, PTRF serves as a promising biomarker in both tumor samples and serum exosomes, thus facilitating the detection of glioma and potentially serving as a therapeutic target for glioblastoma multiforme.
Keywords: Extracellular vesicle; GBM; PTRF/Cavin1; exosome.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

