A highly adaptive microbiome-based association test for survival traits
- PMID: 29558893
- PMCID: PMC5859547
- DOI: 10.1186/s12864-018-4599-8
A highly adaptive microbiome-based association test for survival traits
Abstract
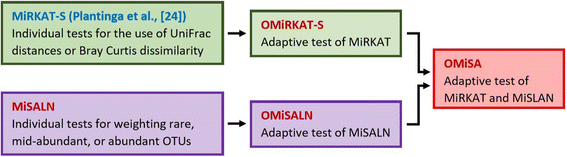
Background: There has been increasing interest in discovering microbial taxa that are associated with human health or disease, gathering momentum through the advances in next-generation sequencing technologies. Investigators have also increasingly employed prospective study designs to survey survival (i.e., time-to-event) outcomes, but current item-by-item statistical methods have limitations due to the unknown true association pattern. Here, we propose a new adaptive microbiome-based association test for survival outcomes, namely, optimal microbiome-based survival analysis (OMiSA). OMiSA approximates to the most powerful association test in two domains: 1) microbiome-based survival analysis using linear and non-linear bases of OTUs (MiSALN) which weighs rare, mid-abundant, and abundant OTUs, respectively, and 2) microbiome regression-based kernel association test for survival traits (MiRKAT-S) which incorporates different distance metrics (e.g., unique fraction (UniFrac) distance and Bray-Curtis dissimilarity), respectively.
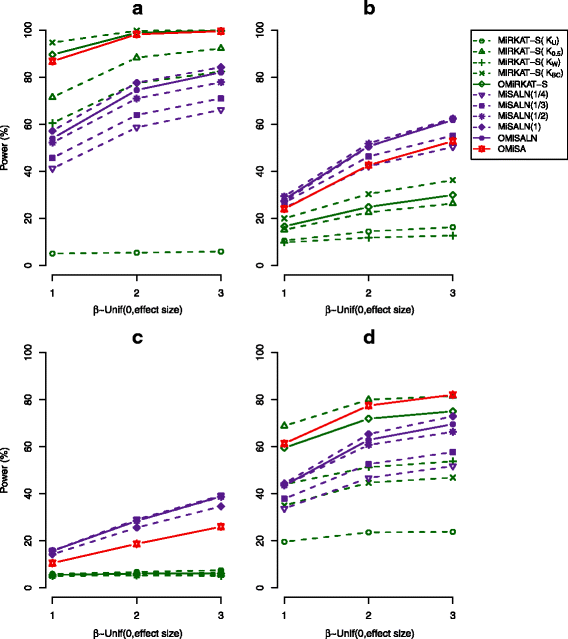
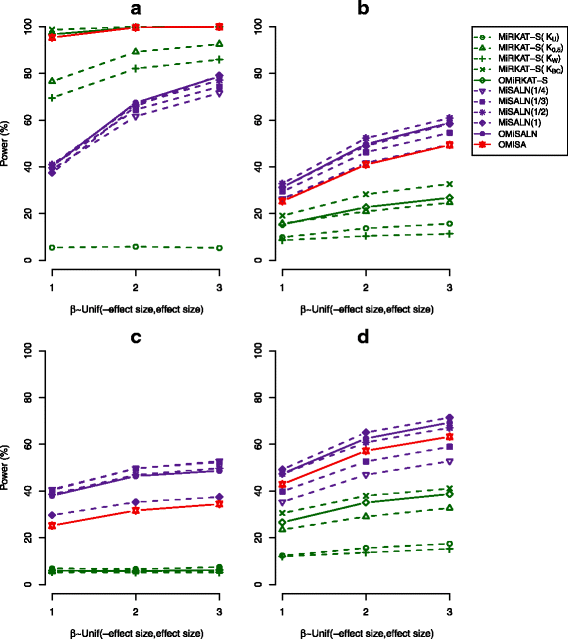
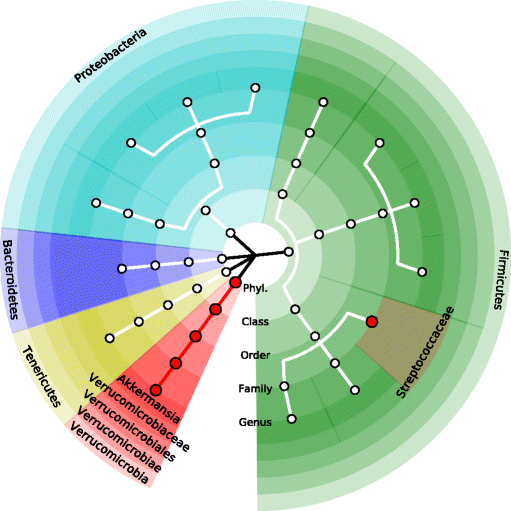
Results: We illustrate that OMiSA powerfully discovers microbial taxa whether their underlying associated lineages are rare or abundant and phylogenetically related or not. OMiSA is a semi-parametric method based on a variance-component score test and a re-sampling method; hence, it is free from any distributional assumption on the effect of microbial composition and advantageous to robustly control type I error rates. Our extensive simulations demonstrate the highly robust performance of OMiSA. We also present the use of OMiSA with real data applications.
Conclusions: OMiSA is attractive in practice as the true association pattern is unpredictable in advance and, for survival outcomes, no adaptive microbiome-based association test is currently available.
Keywords: Community-level association test; High-dimensional compositional data analysis; Microbial group analysis; Microbiome-based association test; Microbiome-based survival analysis; Phylogenetic tree.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable. All utilized data sets are publicly and freely available which do not require any ethics approval and consent to participate.
Consent for publication
Not applicable. All utilized data sets are publicly and freely available which do not require any consent for publication.
Competing interests
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

