Uninfected Bystander Cells Impact the Measurement of HIV-Specific Antibody-Dependent Cellular Cytotoxicity Responses
- PMID: 29559570
- PMCID: PMC5874913
- DOI: 10.1128/mBio.00358-18
Uninfected Bystander Cells Impact the Measurement of HIV-Specific Antibody-Dependent Cellular Cytotoxicity Responses
Abstract
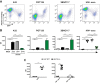
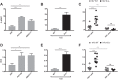
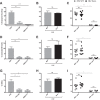
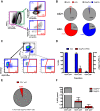
The conformation of the HIV-1 envelope glycoprotein (Env) substantially impacts antibody recognition and antibody-dependent cellular cytotoxicity (ADCC) responses. In the absence of the CD4 receptor at the cell surface, primary Envs sample a "closed" conformation that occludes CD4-induced (CD4i) epitopes. The virus controls CD4 expression through the actions of Nef and Vpu accessory proteins, thus protecting infected cells from ADCC responses. However, gp120 shed from infected cells can bind to CD4 present on uninfected bystander cells, sensitizing them to ADCC mediated by CD4i antibodies (Abs). Therefore, we hypothesized that these bystander cells could impact the interpretation of ADCC measurements. To investigate this, we evaluated the ability of antibodies to CD4i epitopes and broadly neutralizing Abs (bNAbs) to mediate ADCC measured by five ADCC assays commonly used in the field. Our results indicate that the uninfected bystander cells coated with gp120 are efficiently recognized by the CD4i ligands but not the bNabs. Consequently, the uninfected bystander cells substantially affect in vitro measurements made with ADCC assays that fail to identify responses against infected versus uninfected cells. Moreover, using an mRNA flow technique that detects productively infected cells, we found that the vast majority of HIV-1-infected cells in in vitro cultures or ex vivo samples from HIV-1-infected individuals are CD4 negative and therefore do not expose significant levels of CD4i epitopes. Altogether, our results indicate that ADCC assays unable to differentiate responses against infected versus uninfected cells overestimate responses mediated by CD4i ligands.IMPORTANCE Emerging evidence supports a role for antibody-dependent cellular cytotoxicity (ADCC) in protection against HIV-1 transmission and disease progression. However, there are conflicting reports regarding the ability of nonneutralizing antibodies targeting CD4-inducible (CD4i) Env epitopes to mediate ADCC. Here, we performed a side-by-side comparison of different methods currently being used in the field to measure ADCC responses to HIV-1. We found that assays which are unable to differentiate virus-infected from uninfected cells greatly overestimate ADCC responses mediated by antibodies to CD4i epitopes and underestimate responses mediated by broadly neutralizing antibodies (bNAbs). Our results strongly argue for the use of assays that measure ADCC against HIV-1-infected cells expressing physiologically relevant conformations of Env to evaluate correlates of protection in vaccine trials.
Keywords: A32; ADCC; ADCC assay; CD4i Abs; Env; HIV-1; RFADCC; bNAbs; granzyme B assay; luciferase assay; uninfected bystander.
Copyright © 2018 Richard et al.
Figures





Similar articles
-
Antibody-Dependent Cellular Cytotoxicity-Competent Antibodies against HIV-1-Infected Cells in Plasma from HIV-Infected Subjects.mBio. 2019 Dec 17;10(6):e02690-19. doi: 10.1128/mBio.02690-19. mBio. 2019. PMID: 31848282 Free PMC article.
-
CD4 downregulation precedes Env expression and protects HIV-1-infected cells from ADCC mediated by non-neutralizing antibodies.mBio. 2024 Nov 13;15(11):e0182724. doi: 10.1128/mbio.01827-24. Epub 2024 Oct 7. mBio. 2024. PMID: 39373535 Free PMC article.
-
Incomplete Downregulation of CD4 Expression Affects HIV-1 Env Conformation and Antibody-Dependent Cellular Cytotoxicity Responses.J Virol. 2018 Jun 13;92(13):e00484-18. doi: 10.1128/JVI.00484-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29669829 Free PMC article.
-
Impact of HIV-1 Envelope Conformation on ADCC Responses.Trends Microbiol. 2018 Apr;26(4):253-265. doi: 10.1016/j.tim.2017.10.007. Epub 2017 Nov 20. Trends Microbiol. 2018. PMID: 29162391 Review.
-
Unlocking HIV-1 Env: implications for antibody attack.AIDS Res Ther. 2017 Sep 12;14(1):42. doi: 10.1186/s12981-017-0168-5. AIDS Res Ther. 2017. PMID: 28893275 Free PMC article. Review.
Cited by
-
Temsavir blocks the immunomodulatory activities of HIV-1 soluble gp120.Cell Chem Biol. 2023 May 18;30(5):540-552.e6. doi: 10.1016/j.chembiol.2023.03.003. Epub 2023 Mar 22. Cell Chem Biol. 2023. PMID: 36958337 Free PMC article.
-
Pre-existing infant antibody-dependent cellular cytotoxicity associates with reduced HIV-1 acquisition and lower morbidity.Cell Rep Med. 2021 Oct 19;2(10):100412. doi: 10.1016/j.xcrm.2021.100412. eCollection 2021 Oct 19. Cell Rep Med. 2021. PMID: 34755132 Free PMC article.
-
RNA Flow Cytometry for the Study of T Cell Metabolism.Int J Mol Sci. 2021 Apr 9;22(8):3906. doi: 10.3390/ijms22083906. Int J Mol Sci. 2021. PMID: 33918901 Free PMC article. Review.
-
Enhancement of Antibody-Dependent Cellular Cytotoxicity and Phagocytosis in Anti-HIV-1 Human-Bovine Chimeric Broadly Neutralizing Antibodies.J Virol. 2021 Jun 10;95(13):e0021921. doi: 10.1128/JVI.00219-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853957 Free PMC article.
-
A multiclade env-gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques.Nat Med. 2021 Dec;27(12):2234-2245. doi: 10.1038/s41591-021-01574-5. Epub 2021 Dec 9. Nat Med. 2021. PMID: 34887575
References
-
- Veillette M, Coutu M, Richard J, Batraville LA, Dagher O, Bernard N, Tremblay C, Kaufmann DE, Roger M, Finzi A. 2015. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J Virol 89:545–551. doi:10.1128/JVI.02868-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UM1 AI126620/AI/NIAID NIH HHS/United States
- R37 AI095098/AI/NIAID NIH HHS/United States
- UM1 AI100645/AI/NIAID NIH HHS/United States
- UM1 AI100663/AI/NIAID NIH HHS/United States
- R01 AI095098/AI/NIAID NIH HHS/United States
- R01 AI114266/AI/NIAID NIH HHS/United States
- R01 AI121135/AI/NIAID NIH HHS/United States
- P01 AI131251/AI/NIAID NIH HHS/United States
- R01 AI098485/AI/NIAID NIH HHS/United States
- R37 AI055332/AI/NIAID NIH HHS/United States
- P51 OD011106/OD/NIH HHS/United States
- R01 AI055332/AI/NIAID NIH HHS/United States
- R01 AI129769/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

