The genomic and functional landscapes of developmental plasticity in the American cockroach
- PMID: 29559629
- PMCID: PMC5861062
- DOI: 10.1038/s41467-018-03281-1
The genomic and functional landscapes of developmental plasticity in the American cockroach
Abstract
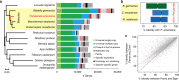
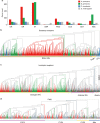
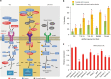
Many cockroach species have adapted to urban environments, and some have been serious pests of public health in the tropics and subtropics. Here, we present the 3.38-Gb genome and a consensus gene set of the American cockroach, Periplaneta americana. We report insights from both genomic and functional investigations into the underlying basis of its adaptation to urban environments and developmental plasticity. In comparison with other insects, expansions of gene families in P. americana exist for most core gene families likely associated with environmental adaptation, such as chemoreception and detoxification. Multiple pathways regulating metamorphic development are well conserved, and RNAi experiments inform on key roles of 20-hydroxyecdysone, juvenile hormone, insulin, and decapentaplegic signals in regulating plasticity. Our analyses reveal a high level of sequence identity in genes between the American cockroach and two termite species, advancing it as a valuable model to study the evolutionary relationships between cockroaches and termites.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Improved genome assembly provides new insights into the environmental adaptation of the American cockroach, Periplaneta americana.Arch Insect Biochem Physiol. 2022 Dec;111(4):e21956. doi: 10.1002/arch.21956. Epub 2022 Aug 7. Arch Insect Biochem Physiol. 2022. PMID: 35933728
-
The hormonal pathway controlling cell death during metamorphosis in a hemimetabolous insect.Dev Biol. 2010 Oct 1;346(1):150-60. doi: 10.1016/j.ydbio.2010.07.012. Epub 2010 Jul 16. Dev Biol. 2010. PMID: 20638378
-
Complete mitochondrial genomes of two cockroaches, Blattella germanica and Periplaneta americana, and the phylogenetic position of termites.Curr Genet. 2012 Apr;58(2):65-77. doi: 10.1007/s00294-012-0365-7. Epub 2012 Feb 7. Curr Genet. 2012. PMID: 22311390
-
Nutrition- and hormone-controlled developmental plasticity in Blattodea.Curr Opin Insect Sci. 2023 Dec;60:101128. doi: 10.1016/j.cois.2023.101128. Epub 2023 Oct 6. Curr Opin Insect Sci. 2023. PMID: 37806339 Review.
-
Integrated genomics approaches in evolutionary and ecological endocrinology.Adv Exp Med Biol. 2014;781:299-319. doi: 10.1007/978-94-007-7347-9_15. Adv Exp Med Biol. 2014. PMID: 24277306 Review.
Cited by
-
The Draft Genome Dataset of the Asian Cricket Teleogryllus occipitalis for Molecular Research Toward Entomophagy.Front Genet. 2020 May 8;11:470. doi: 10.3389/fgene.2020.00470. eCollection 2020. Front Genet. 2020. PMID: 32457806 Free PMC article. No abstract available.
-
Hox-determined appendage regeneration restores cockroach courtship rituals.Sci China Life Sci. 2025 Mar;68(3):874-876. doi: 10.1007/s11427-024-2767-3. Epub 2024 Dec 5. Sci China Life Sci. 2025. PMID: 39643834 No abstract available.
-
The Bear Giant-Skipper genome suggests genetic adaptations to living inside yucca roots.Mol Genet Genomics. 2019 Feb;294(1):211-226. doi: 10.1007/s00438-018-1494-6. Epub 2018 Oct 6. Mol Genet Genomics. 2019. PMID: 30293092 Free PMC article.
-
Of Cockroaches and Symbionts: Recent Advances in the Characterization of the Relationship between Blattella germanica and Its Dual Symbiotic System.Life (Basel). 2022 Feb 15;12(2):290. doi: 10.3390/life12020290. Life (Basel). 2022. PMID: 35207577 Free PMC article. Review.
-
Life-History Traits from Embryonic Development to Reproduction in the American Cockroach.Insects. 2022 Jun 16;13(6):551. doi: 10.3390/insects13060551. Insects. 2022. PMID: 35735888 Free PMC article.
References
-
- Bell, W. J. & Adiyodi, K. The American Cockroach (Springer Science & Business Media, New York, 1982).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

