Characterization of a murine model of non-lethal, symptomatic dengue virus infection
- PMID: 29559699
- PMCID: PMC5861036
- DOI: 10.1038/s41598-018-22618-w
Characterization of a murine model of non-lethal, symptomatic dengue virus infection
Abstract
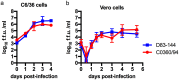
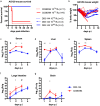
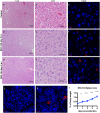
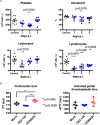
The mosquito-borne disease dengue is caused by four serologically- and genetically-related viruses, termed DENV-1 to DENV-4. Historical setbacks due to lack of human-like mouse models of dengue were partially remedied with characterization of lethal DENV-2 infection in immunocompromised AG129 mice (deficient in IFN-α/β/γ receptors). Recently, our group established lethal AG129 mouse infection models of DENV-1, DENV-3, and DENV-4 using human isolates. Here we compare a non-lethal, disseminated model of DENV-3 infection using strain D83-144 to that of the lethal outcome following infection by strain C0360/94. Both strains belong to DENV-3 genotype II and differ by only 13 amino acids. Intraperitoneal inoculation of AG129 mice with strain D83-144 led to clinical signs of dengue infection, such as cytokine induction, thrombocytopenia, and systemic infection. However, C0360/94 infection led to features of severe human dengue, including coagulopathy and lethal outcome, whereas D83-144 infection does not. This study is the first to investigate a low passage, non-mouse lethal strain in AG129 mice and demonstrates that D83-144 infection induces milder features of human dengue than those induced by lethal C0360/94 infection. The results suggest that the AG129 mouse model has applications to investigate factors associated with mild or severe disease.
Conflict of interest statement
The authors declare no competing interests.
Figures

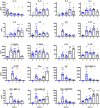

Similar articles
-
A lethal murine infection model for dengue virus 3 in AG129 mice deficient in type I and II interferon receptors leads to systemic disease.J Virol. 2015 Jan 15;89(2):1254-66. doi: 10.1128/JVI.01320-14. Epub 2014 Nov 12. J Virol. 2015. PMID: 25392217 Free PMC article.
-
Characterization of lethal dengue virus type 4 (DENV-4) TVP-376 infection in mice lacking both IFN-α/β and IFN-γ receptors (AG129) and comparison with the DENV-2 AG129 mouse model.J Gen Virol. 2015 Oct;96(10):3035-3048. doi: 10.1099/jgv.0.000246. Epub 2015 Jul 14. J Gen Virol. 2015. PMID: 26296350 Free PMC article.
-
A lethal model of disseminated dengue virus type 1 infection in AG129 mice.J Gen Virol. 2017 Oct;98(10):2507-2519. doi: 10.1099/jgv.0.000923. Epub 2017 Sep 27. J Gen Virol. 2017. PMID: 28949904 Free PMC article.
-
Mouse models of dengue virus infection for vaccine testing.Vaccine. 2015 Dec 10;33(50):7051-60. doi: 10.1016/j.vaccine.2015.09.112. Epub 2015 Oct 23. Vaccine. 2015. PMID: 26478201 Free PMC article. Review.
-
Influence of antibodies and T cells on dengue disease outcome: insights from interferon receptor-deficient mouse models.Curr Opin Virol. 2015 Aug;13:61-6. doi: 10.1016/j.coviro.2015.04.007. Epub 2015 May 23. Curr Opin Virol. 2015. PMID: 26001278 Review.
Cited by
-
Evaluation of Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus Mosquitoes Competence to Oropouche virus Infection.Viruses. 2021 Apr 25;13(5):755. doi: 10.3390/v13050755. Viruses. 2021. PMID: 33923055 Free PMC article.
-
Robust Plasma Cell Response to Skin-Inoculated Dengue Virus in Mice.J Immunol Res. 2021 Apr 26;2021:5511841. doi: 10.1155/2021/5511841. eCollection 2021. J Immunol Res. 2021. PMID: 33997054 Free PMC article.
-
Evaluating Dengue Virus Pathogenesis in Mice and Humans by Histological and Immunohistochemistry Approaches.Methods Mol Biol. 2022;2409:259-269. doi: 10.1007/978-1-0716-1879-0_18. Methods Mol Biol. 2022. PMID: 34709648
-
Identification of Dengue Virus Serotype 3 Specific Antigenic Sites Targeted by Neutralizing Human Antibodies.Cell Host Microbe. 2020 May 13;27(5):710-724.e7. doi: 10.1016/j.chom.2020.04.007. Cell Host Microbe. 2020. PMID: 32407709 Free PMC article.
-
Use of Animal Models in Studying Roles of Antibodies and Their Secretion Cells in Dengue Vaccine Development.Viruses. 2020 Nov 5;12(11):1261. doi: 10.3390/v12111261. Viruses. 2020. PMID: 33167518 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

