Molecular Networks of Postia placenta Involved in Degradation of Lignocellulosic Biomass Revealed from Metadata Analysis of Open Access Gene Expression Data
- PMID: 29559843
- PMCID: PMC5859471
- DOI: 10.7150/ijbs.22868
Molecular Networks of Postia placenta Involved in Degradation of Lignocellulosic Biomass Revealed from Metadata Analysis of Open Access Gene Expression Data
Abstract
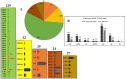
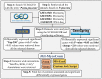
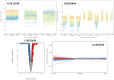
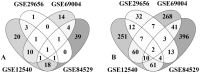
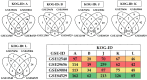
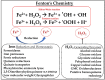
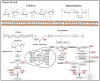
To understand the common gene expression patterns employed by P. placenta during lignocellulose degradation, we have retrieved genome wide transcriptome datasets from NCBI GEO database and analyzed using customized analysis pipeline. We have retrieved the top differentially expressed genes and compared the common significant genes among two different growth conditions. Genes encoding for cellulolytic (GH1, GH3, GH5, GH12, GH16, GH45) and hemicellulolytic (GH10, GH27, GH31, GH35, GH47, GH51, GH55, GH78, GH95) glycoside hydrolase classes were commonly up regulated among all the datasets. Fenton's reaction enzymes (iron homeostasis, reduction, hydrogen peroxide generation) were significantly expressed among all the datasets under lignocellulolytic conditions. Due to the evolutionary loss of genes coding for various lignocellulolytic enzymes (including several cellulases), P. placenta employs hemicellulolytic glycoside hydrolases and Fenton's reactions for the rapid depolymerization of plant cell wall components. Different classes of enzymes involved in aromatic compound degradation, stress responsive and detoxification mechanisms (cytochrome P450 monoxygenases) were found highly expressed in complex plant biomass substrates. We have reported the genome wide expression patterns of genes coding for information, storage and processing (KOG), tentative and predicted molecular networks involved in cellulose, hemicellulose degradation and list of significant protein-ID's commonly expressed among different lignocellulolytic growth conditions.
Keywords: Brown-rot decay; Fenton's reaction; Gene expression; Lignocellulose; NCBI-Gene Expression Omnibus (GEO); Postia placenta.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


Similar articles
-
Systematic metadata analysis of brown rot fungi gene expression data reveals the genes involved in Fenton's reaction and wood decay process.Mycology. 2019 Dec 24;11(1):22-37. doi: 10.1080/21501203.2019.1703052. eCollection 2020. Mycology. 2019. PMID: 32128279 Free PMC article.
-
Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion.Proc Natl Acad Sci U S A. 2009 Feb 10;106(6):1954-9. doi: 10.1073/pnas.0809575106. Epub 2009 Feb 4. Proc Natl Acad Sci U S A. 2009. PMID: 19193860 Free PMC article.
-
Metadata Analysis of Phanerochaete chrysosporium Gene Expression Data Identified Common CAZymes Encoding Gene Expression Profiles Involved in Cellulose and Hemicellulose Degradation.Int J Biol Sci. 2017 Jan 1;13(1):85-99. doi: 10.7150/ijbs.17390. eCollection 2017. Int J Biol Sci. 2017. PMID: 28123349 Free PMC article.
-
Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates.Enzyme Microb Technol. 2017 Aug;103:1-11. doi: 10.1016/j.enzmictec.2017.04.007. Epub 2017 Apr 25. Enzyme Microb Technol. 2017. PMID: 28554379 Review.
-
Recent Developments in Using Advanced Sequencing Technologies for the Genomic Studies of Lignin and Cellulose Degrading Microorganisms.Int J Biol Sci. 2016 Jan 1;12(2):156-71. doi: 10.7150/ijbs.13537. eCollection 2016. Int J Biol Sci. 2016. PMID: 26884714 Free PMC article. Review.
Cited by
-
Metagenomic and physicochemical profiling reveal microbial functions in pit mud for Jiang-Nong Jianxiang Baijiu fermentation.BMC Microbiol. 2025 Apr 2;25(1):190. doi: 10.1186/s12866-025-03884-x. BMC Microbiol. 2025. PMID: 40175903 Free PMC article.
-
Parascedosporium putredinis NO1 tailors its secretome for different lignocellulosic substrates.Microbiol Spectr. 2024 Jul 2;12(7):e0394323. doi: 10.1128/spectrum.03943-23. Epub 2024 May 17. Microbiol Spectr. 2024. PMID: 38757984 Free PMC article.
-
Oxidative stress facilitates infection of the unicellular alga Haematococcus pluvialis by the fungus Paraphysoderma sedebokerense.Biotechnol Biofuels Bioprod. 2022 May 20;15(1):56. doi: 10.1186/s13068-022-02140-y. Biotechnol Biofuels Bioprod. 2022. PMID: 35596207 Free PMC article.
-
Systematic metadata analysis of brown rot fungi gene expression data reveals the genes involved in Fenton's reaction and wood decay process.Mycology. 2019 Dec 24;11(1):22-37. doi: 10.1080/21501203.2019.1703052. eCollection 2020. Mycology. 2019. PMID: 32128279 Free PMC article.
-
Metadata Analysis Approaches for Understanding and Improving the Functional Involvement of Rumen Microbial Consortium in Digestion and Metabolism of Plant Biomass.J Genomics. 2019 Apr 2;7:31-45. doi: 10.7150/jgen.32164. eCollection 2019. J Genomics. 2019. PMID: 31001361 Free PMC article. Review.
References
-
- Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP. et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proceedings of the National Academy of Sciences. 2009;106:1954–9. - PMC - PubMed
-
- Binder M, Hibbett DS, Larsson KH, Larsson E, Langer E, Langer G. The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes) Systematics and Biodiversity. 2005;3:113–57.
-
- Hibbett DS, Donoghue MJ. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Systematic biology; 2001. pp. 215–42. - PubMed
-
- Arantes V, Goodell B. Current understanding of brown-rot fungal biodegradation mechanisms: a review. Deterioration and protection of sustainable biomaterials. 2014;1158:3–21.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

