RNAi-Mediated Downregulation of Inositol Pentakisphosphate Kinase (IPK1) in Wheat Grains Decreases Phytic Acid Levels and Increases Fe and Zn Accumulation
- PMID: 29559984
- PMCID: PMC5845732
- DOI: 10.3389/fpls.2018.00259
RNAi-Mediated Downregulation of Inositol Pentakisphosphate Kinase (IPK1) in Wheat Grains Decreases Phytic Acid Levels and Increases Fe and Zn Accumulation
Abstract
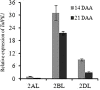
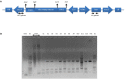
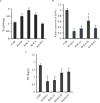
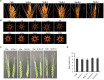
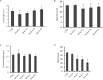
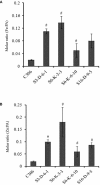
Enhancement of micronutrient bioavailability is crucial to address the malnutrition in the developing countries. Various approaches employed to address the micronutrient bioavailability are showing promising signs, especially in cereal crops. Phytic acid (PA) is considered as a major antinutrient due to its ability to chelate important micronutrients and thereby restricting their bioavailability. Therefore, manipulating PA biosynthesis pathway has largely been explored to overcome the pleiotropic effect in different crop species. Recently, we reported that functional wheat inositol pentakisphosphate kinase (TaIPK1) is involved in PA biosynthesis, however, the functional roles of the IPK1 gene in wheat remains elusive. In this study, RNAi-mediated gene silencing was performed for IPK1 transcripts in hexaploid wheat. Four non-segregating RNAi lines of wheat were selected for detailed study (S3-D-6-1; S6-K-3-3; S6-K-6-10 and S16-D-9-5). Homozygous transgenic RNAi lines at T4 seeds with a decreased transcript of TaIPK1 showed 28-56% reduction of the PA. Silencing of IPK1 also resulted in increased free phosphate in mature grains. Although, no phenotypic changes in the spike was observed but, lowering of grain PA resulted in the reduced number of seeds per spikelet. The lowering of grain PA was also accompanied by a significant increase in iron (Fe) and zinc (Zn) content, thereby enhancing their molar ratios (Zn:PA and Fe:PA). Overall, this work suggests that IPK1 is a promising candidate for employing genome editing tools to address the mineral accumulation in wheat grains.
Keywords: Triticum aestivum; gene silencing; inositol pentakisphosphate kinase; phytic acid; wheat transformation.
Figures
References
-
- Ames B. N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8 115–118. 10.1016/0076-6879(66)08014-5 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

