Directed evolution of GFP with non-natural amino acids identifies residues for augmenting and photoswitching fluorescence
- PMID: 29560203
- PMCID: PMC5811120
- DOI: 10.1039/c4sc02827a
Directed evolution of GFP with non-natural amino acids identifies residues for augmenting and photoswitching fluorescence
Abstract
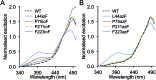
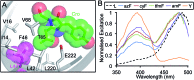
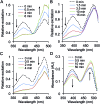
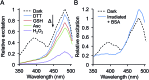
Genetic code reprogramming allows proteins to sample new chemistry through the defined and targeted introduction of non-natural amino acids (nAAs). Many useful nAAs are derivatives of the natural aromatic amino acid tyrosine, with the para OH group replaced with useful but often bulkier substituents. Extending residue sampling by directed evolution identified positions in Green Fluorescent Protein tolerant to aromatic nAAs, including identification of novel sites that modulate fluorescence. Replacement of the buried L44 residue by photosensitive p-azidophenylalanine (azF) conferred environmentally sensitive photoswitching. In silico modelling of the L44azF dark state provided an insight into the mechanism of action through modulation of the hydrogen bonding network surrounding the chromophore. Targeted mutagenesis of T203 with aromatic nAAs to introduce π-stacking with the chromophore successfully generated red shifted versions of GFP. Incorporation of azF at residue 203 conferred high photosensitivity on sfGFP with even ambient light mediating a functional switch. Thus, engineering proteins with non-natural aromatic amino acids by surveying a wide residue set can introduce new and beneficial properties into a protein through the sampling of non-intuitive mutations. Coupled with retrospective in silico modelling, this will facilitate both our understanding of the impact of nAAs on protein structure and function, and future design endeavours.
Figures

Similar articles
-
In-frame amber stop codon replacement mutagenesis for the directed evolution of proteins containing non-canonical amino acids: identification of residues open to bio-orthogonal modification.PLoS One. 2015 May 26;10(5):e0127504. doi: 10.1371/journal.pone.0127504. eCollection 2015. PLoS One. 2015. PMID: 26011713 Free PMC article.
-
Protein evolution via amino acid and codon elimination.PLoS One. 2010 Apr 26;5(4):e10104. doi: 10.1371/journal.pone.0010104. PLoS One. 2010. PMID: 20436666 Free PMC article.
-
The 1.7 A crystal structure of Dronpa: a photoswitchable green fluorescent protein.J Mol Biol. 2006 Nov 24;364(2):213-24. doi: 10.1016/j.jmb.2006.08.089. Epub 2006 Sep 3. J Mol Biol. 2006. PMID: 17010376
-
Photoswitching of E222Q GFP mutants: "concerted" mechanism of chromophore isomerization and protonation.Photochem Photobiol Sci. 2010 Oct 28;9(10):1307-19. doi: 10.1039/c0pp00189a. Epub 2010 Sep 21. Photochem Photobiol Sci. 2010. PMID: 20859582 Review.
-
Function and therapeutic potential of N-acyl amino acids.Chem Phys Lipids. 2021 Sep;239:105114. doi: 10.1016/j.chemphyslip.2021.105114. Epub 2021 Jul 2. Chem Phys Lipids. 2021. PMID: 34217720 Review.
Cited by
-
Stalling chromophore synthesis of the fluorescent protein Venus reveals the molecular basis of the final oxidation step.Chem Sci. 2021 Mar 31;12(22):7735-7745. doi: 10.1039/d0sc06693a. Chem Sci. 2021. PMID: 34168826 Free PMC article.
-
A general strategy to red-shift green fluorescent protein-based biosensors.Nat Chem Biol. 2020 Dec;16(12):1434-1439. doi: 10.1038/s41589-020-0641-7. Epub 2020 Sep 14. Nat Chem Biol. 2020. PMID: 32929278 Free PMC article.
-
Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.J Am Chem Soc. 2019 Sep 25;141(38):15250-15265. doi: 10.1021/jacs.9b07152. Epub 2019 Sep 11. J Am Chem Soc. 2019. PMID: 31450887 Free PMC article.
-
Functional modulation and directed assembly of an enzyme through designed non-natural post-translation modification.Chem Sci. 2015 Jul 15;6(7):3712-3717. doi: 10.1039/c4sc03900a. Epub 2015 Mar 31. Chem Sci. 2015. PMID: 28706718 Free PMC article.
-
Designed Artificial Protein Heterodimers With Coupled Functions Constructed Using Bio-Orthogonal Chemistry.Front Chem. 2021 Aug 4;9:733550. doi: 10.3389/fchem.2021.733550. eCollection 2021. Front Chem. 2021. PMID: 34422774 Free PMC article.
References
-
- Zhang W. H., Otting G., Jackson C. J. Curr. Opin. Struct. Biol. 2013;23:581–587. - PubMed
-
- Antonczak A. K., Morris J., Tippmann E. M. Curr. Opin. Struct. Biol. 2011;21:481–487. - PubMed
-
- Reddington S., Watson P., Rizkallah P., Tippmann E., Jones D. D. Biochem. Soc. Trans. 2013;41:1177–1182. - PubMed
-
- Wang L., Magliery T., Liu D., Schultz P. J. Am. Chem. Soc. 2000;122:5010–5011.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

