Natural variation of chronological aging in the Saccharomyces cerevisiae species reveals diet-dependent mechanisms of life span control
- PMID: 29560271
- PMCID: PMC5845861
- DOI: 10.1038/s41514-018-0022-6
Natural variation of chronological aging in the Saccharomyces cerevisiae species reveals diet-dependent mechanisms of life span control
Abstract
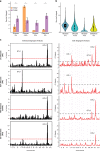
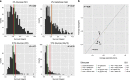
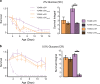
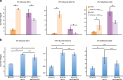
Aging is a complex trait of broad scientific interest, especially because of its intrinsic link with common human diseases. Pioneering work on aging-related mechanisms has been made in Saccharomyces cerevisiae, mainly through the use of deletion collections isogenic to the S288c reference strain. In this study, using a recently published high-throughput approach, we quantified chronological life span (CLS) within a collection of 58 natural strains across seven different conditions. We observed a broad aging variability suggesting the implication of diverse genetic and environmental factors in chronological aging control. Two major Quantitative Trait Loci (QTLs) were identified within a biparental population obtained by crossing two natural isolates with contrasting aging behavior. Detection of these QTLs was dependent upon the nature and concentration of the carbon sources available for growth. In the first QTL, the RIM15 gene was identified as major regulator of aging under low glucose condition, lending further support to the importance of nutrient-sensing pathways in longevity control under calorie restriction. In the second QTL, we could show that the SER1 gene, encoding a conserved aminotransferase of the serine synthesis pathway not previously linked to aging, is causally associated with CLS regulation, especially under high glucose condition. These findings hint toward a new mechanism of life span control involving a trade-off between serine synthesis and aging, most likely through modulation of acetate and trehalose metabolism. More generally it shows that genetic linkage studies across natural strains represent a promising strategy to further unravel the molecular basis of aging.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Gene-nutrient interaction markedly influences yeast chronological lifespan.Exp Gerontol. 2016 Dec 15;86:113-123. doi: 10.1016/j.exger.2016.04.012. Epub 2016 Apr 25. Exp Gerontol. 2016. PMID: 27125759 Free PMC article.
-
Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins.Aging Cell. 2007 Oct;6(5):649-62. doi: 10.1111/j.1474-9726.2007.00326.x. Epub 2007 Aug 15. Aging Cell. 2007. PMID: 17711561
-
Nitrogen and carbon source balance determines longevity, independently of fermentative or respiratory metabolism in the yeast Saccharomyces cerevisiae.Oncotarget. 2016 Apr 26;7(17):23033-42. doi: 10.18632/oncotarget.8656. Oncotarget. 2016. PMID: 27072582 Free PMC article.
-
The chronological life span of Saccharomyces cerevisiae.Methods Mol Biol. 2007;371:89-95. doi: 10.1007/978-1-59745-361-5_8. Methods Mol Biol. 2007. PMID: 17634576 Review.
-
The nature of quantitative genetic variation for Drosophila longevity.Mech Ageing Dev. 2002 Jan;123(2-3):95-104. doi: 10.1016/s0047-6374(01)00330-x. Mech Ageing Dev. 2002. PMID: 11718804 Review.
Cited by
-
An Optimized Competitive-Aging Method Reveals Gene-Drug Interactions Underlying the Chronological Lifespan of Saccharomyces cerevisiae.Front Genet. 2020 May 14;11:468. doi: 10.3389/fgene.2020.00468. eCollection 2020. Front Genet. 2020. PMID: 32477409 Free PMC article.
-
Exploring the anti-aging potential of natural products and plant extracts in budding yeast Saccharomyces cerevisiae: A review.F1000Res. 2024 Dec 17;12:1265. doi: 10.12688/f1000research.141669.2. eCollection 2023. F1000Res. 2024. PMID: 39822944 Free PMC article. Review.
-
Identification of novel genes involved in neutral lipid storage by quantitative trait loci analysis of Saccharomyces cerevisiae.BMC Genomics. 2021 Feb 9;22(1):110. doi: 10.1186/s12864-021-07417-4. BMC Genomics. 2021. PMID: 33563210 Free PMC article.
-
The SESAME complex regulates cell senescence through the generation of acetyl-CoA.Nat Metab. 2021 Jul;3(7):983-1000. doi: 10.1038/s42255-021-00412-9. Epub 2021 Jun 28. Nat Metab. 2021. PMID: 34183849
-
A prion accelerates proliferation at the expense of lifespan.Elife. 2021 Sep 15;10:e60917. doi: 10.7554/eLife.60917. Elife. 2021. PMID: 34545808 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

