Large-scale computational drug repositioning to find treatments for rare diseases
- PMID: 29560273
- PMCID: PMC5847522
- DOI: 10.1038/s41540-018-0050-7
Large-scale computational drug repositioning to find treatments for rare diseases
Abstract
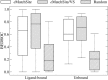
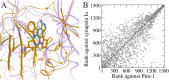
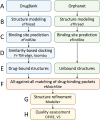
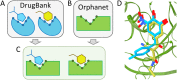
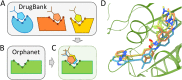
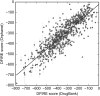
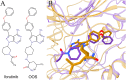
Rare, or orphan, diseases are conditions afflicting a small subset of people in a population. Although these disorders collectively pose significant health care problems, drug companies require government incentives to develop drugs for rare diseases due to extremely limited individual markets. Computer-aided drug repositioning, i.e., finding new indications for existing drugs, is a cheaper and faster alternative to traditional drug discovery offering a promising venue for orphan drug research. Structure-based matching of drug-binding pockets is among the most promising computational techniques to inform drug repositioning. In order to find new targets for known drugs ultimately leading to drug repositioning, we recently developed eMatchSite, a new computer program to compare drug-binding sites. In this study, eMatchSite is combined with virtual screening to systematically explore opportunities to reposition known drugs to proteins associated with rare diseases. The effectiveness of this integrated approach is demonstrated for a kinase inhibitor, which is a confirmed candidate for repositioning to synapsin Ia. The resulting dataset comprises 31,142 putative drug-target complexes linked to 980 orphan diseases. The modeling accuracy is evaluated against the structural data recently released for tyrosine-protein kinase HCK. To illustrate how potential therapeutics for rare diseases can be identified, we discuss a possibility to repurpose a steroidal aromatase inhibitor to treat Niemann-Pick disease type C. Overall, the exhaustive exploration of the drug repositioning space exposes new opportunities to combat orphan diseases with existing drugs. DrugBank/Orphanet repositioning data are freely available to research community at https://osf.io/qdjup/.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
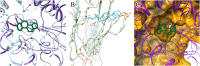
Similar articles
-
eRepo-ORP: Exploring the Opportunity Space to Combat Orphan Diseases with Existing Drugs.J Mol Biol. 2018 Jul 20;430(15):2266-2273. doi: 10.1016/j.jmb.2017.12.001. Epub 2017 Dec 10. J Mol Biol. 2018. PMID: 29237557 Free PMC article.
-
Local Alignment of Ligand Binding Sites in Proteins for Polypharmacology and Drug Repositioning.Methods Mol Biol. 2017;1611:109-122. doi: 10.1007/978-1-4939-7015-5_9. Methods Mol Biol. 2017. PMID: 28451975 Free PMC article.
-
A systems-level analysis of drug-target-disease associations for drug repositioning.Brief Funct Genomics. 2018 Jan 1;17(1):34-41. doi: 10.1093/bfgp/elx015. Brief Funct Genomics. 2018. PMID: 28968683
-
Drug repositioning for orphan diseases.Brief Bioinform. 2011 Jul;12(4):346-56. doi: 10.1093/bib/bbr021. Epub 2011 Apr 18. Brief Bioinform. 2011. PMID: 21504985 Review.
-
Drug repositioning for personalized medicine.Genome Med. 2012 Mar 30;4(3):27. doi: 10.1186/gm326. eCollection 2012. Genome Med. 2012. PMID: 22494857 Free PMC article. Review.
Cited by
-
In silico screening of phenylethanoid glycosides, a class of pharmacologically active compounds as natural inhibitors of SARS-CoV-2 proteases.Comput Struct Biotechnol J. 2023;21:1461-1472. doi: 10.1016/j.csbj.2023.02.020. Epub 2023 Feb 11. Comput Struct Biotechnol J. 2023. PMID: 36817956 Free PMC article.
-
GraphSite: Ligand Binding Site Classification with Deep Graph Learning.Biomolecules. 2022 Jul 29;12(8):1053. doi: 10.3390/biom12081053. Biomolecules. 2022. PMID: 36008947 Free PMC article.
-
Suppression of LPS-Induced Inflammation and Cell Migration by Azelastine through Inhibition of JNK/NF-κB Pathway in BV2 Microglial Cells.Int J Mol Sci. 2021 Aug 23;22(16):9061. doi: 10.3390/ijms22169061. Int J Mol Sci. 2021. PMID: 34445767 Free PMC article.
-
Drug Repositioning for Noonan and LEOPARD Syndromes by Integrating Transcriptomics With a Structure-Based Approach.Front Pharmacol. 2020 Jun 26;11:927. doi: 10.3389/fphar.2020.00927. eCollection 2020. Front Pharmacol. 2020. PMID: 32676024 Free PMC article.
-
Prediction of drug's anatomical therapeutic chemical (ATC) code by constructing biological profiles of ATC codes.BMC Bioinformatics. 2025 Mar 21;26(1):86. doi: 10.1186/s12859-025-06102-7. BMC Bioinformatics. 2025. PMID: 40119265 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

