In silico drug combination discovery for personalized cancer therapy
- PMID: 29560824
- PMCID: PMC5861486
- DOI: 10.1186/s12918-018-0546-1
In silico drug combination discovery for personalized cancer therapy
Abstract
Background: Drug combination therapy, which is considered as an alternative to single drug therapy, can potentially reduce resistance and toxicity, and have synergistic efficacy. As drug combination therapies are widely used in the clinic for hypertension, asthma, and AIDS, they have also been proposed for the treatment of cancer. However, it is difficult to select and experimentally evaluate effective combinations because not only is the number of cancer drug combinations extremely large but also the effectiveness of drug combinations varies depending on the genetic variation of cancer patients. A computational approach that prioritizes the best drug combinations considering the genetic information of a cancer patient is necessary to reduce the search space.
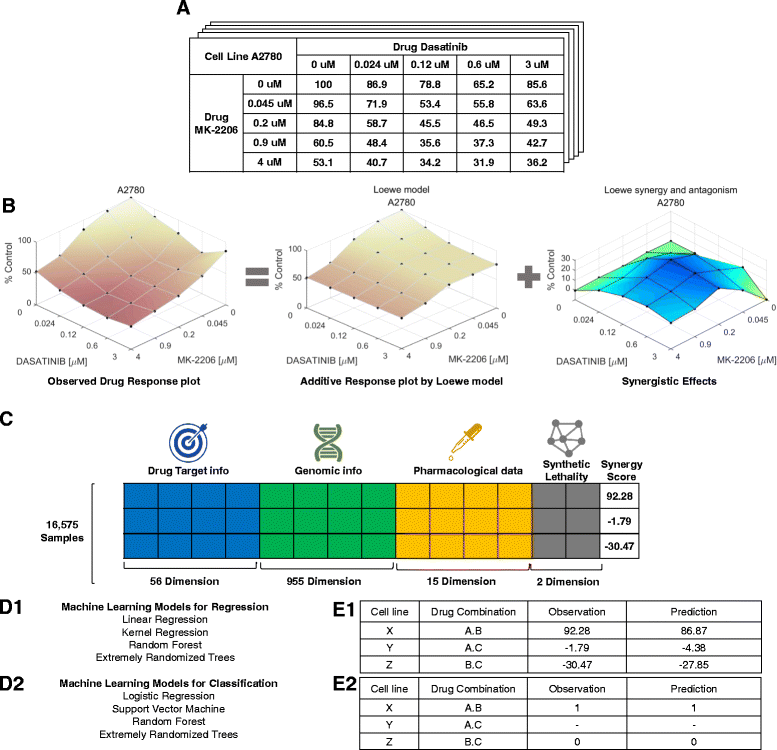
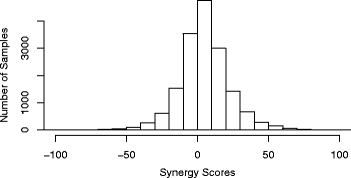
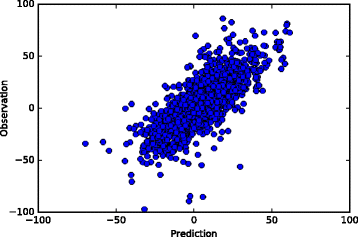
Results: We propose an in-silico method for personalized drug combination therapy discovery. We predict the synergy between two drugs and a cell line using genomic information, targets of drugs, and pharmacological information. We calculate and predict the synergy scores of 583 drug combinations for 31 cancer cell lines. For feature dimension reduction, we select the mutations or expression levels of the genes in cancer-related pathways. We also used various machine learning models. Extremely Randomized Trees (ERT), a tree-based ensemble model, achieved the best performance in the synergy score prediction regression task. The correlation coefficient between the synergy scores predicted by ERT and the actual observations is 0.738. To compare with an existing drug combination synergy classification model, we reformulate the problem as a binary classification problem by thresholding the synergy scores. ERT achieved an F1 score of 0.954 when synergy scores of 20 and -20 were used as the threshold, which is 8.7% higher than that obtained by the state-of-the-art baseline model. Moreover, the model correctly predicts the most synergistic combination, from approximately 100 candidate drug combinations, as the top choice for 15 out of the 31 cell lines. For 28 out of the 31 cell lines, the model predicts the most synergistic combination in the top 10 of approximately 100 candidate drug combinations. Finally, we analyze the results, generate synergistic rules using the features, and validate the rules through the literature survey.
Conclusion: Using various types of genomic information of cancer cell lines, targets of drugs, and pharmacological information, a drug combination synergy prediction pipeline is proposed. The pipeline regresses the synergy level between two drugs and a cell line as well as classifies if there exists synergy or antagonism between them. Discovering new drug combinations by our pipeline may improve personalized cancer therapy.
Keywords: Combination therapy; Synergy prediction; in silico.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Synergy from gene expression and network mining (SynGeNet) method predicts synergistic drug combinations for diverse melanoma genomic subtypes.NPJ Syst Biol Appl. 2019 Feb 26;5:6. doi: 10.1038/s41540-019-0085-4. eCollection 2019. NPJ Syst Biol Appl. 2019. PMID: 30820351 Free PMC article.
-
An integrated framework for identification of effective and synergistic anti-cancer drug combinations.J Bioinform Comput Biol. 2018 Oct;16(5):1850017. doi: 10.1142/S0219720018500178. Epub 2018 Jun 28. J Bioinform Comput Biol. 2018. PMID: 30304987
-
Synergistic Drug Combination Prediction by Integrating Multiomics Data in Deep Learning Models.Methods Mol Biol. 2021;2194:223-238. doi: 10.1007/978-1-0716-0849-4_12. Methods Mol Biol. 2021. PMID: 32926369
-
Discovering Synergistic Drug Combination from a Computational Perspective.Curr Top Med Chem. 2018;18(12):965-974. doi: 10.2174/1568026618666180330141804. Curr Top Med Chem. 2018. PMID: 29600766 Review.
-
You Cannot Have Your Synergy and Efficacy Too.Trends Pharmacol Sci. 2019 Nov;40(11):811-817. doi: 10.1016/j.tips.2019.08.008. Epub 2019 Oct 11. Trends Pharmacol Sci. 2019. PMID: 31610891 Review.
Cited by
-
HTSplotter: An end-to-end data processing, analysis and visualisation tool for chemical and genetic in vitro perturbation screening.PLoS One. 2024 Jan 5;19(1):e0296322. doi: 10.1371/journal.pone.0296322. eCollection 2024. PLoS One. 2024. PMID: 38181013 Free PMC article.
-
8-Hydroxydaidzein Induces Apoptosis and Inhibits AML-Associated Gene Expression in U-937 Cells: Potential Phytochemical for AML Treatment.Biomolecules. 2023 Oct 26;13(11):1575. doi: 10.3390/biom13111575. Biomolecules. 2023. PMID: 38002257 Free PMC article.
-
Boolean modeling of breast cancer signaling pathways uncovers mechanisms of drug synergy.PLoS One. 2024 Feb 23;19(2):e0298788. doi: 10.1371/journal.pone.0298788. eCollection 2024. PLoS One. 2024. PMID: 38394152 Free PMC article.
-
PermuteDDS: a permutable feature fusion network for drug-drug synergy prediction.J Cheminform. 2024 Apr 15;16(1):41. doi: 10.1186/s13321-024-00839-8. J Cheminform. 2024. PMID: 38622663 Free PMC article.
-
The circadian rhythm: an influential soundtrack in the diabetes story.Front Endocrinol (Lausanne). 2023 Jun 27;14:1156757. doi: 10.3389/fendo.2023.1156757. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37441501 Free PMC article. Review.
References
-
- Clinical Development Success Rates 2006-2015. https://www.bio.org/sites/default/files/ClinicalDevelopmentSuccessRates2.... Accessed 30 Oct 2017.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

