Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research
- PMID: 29561359
- PMCID: PMC6008200
- DOI: 10.1097/j.pain.0000000000001217
Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research
Abstract
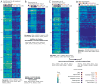
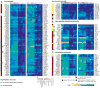
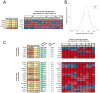
Molecular neurobiological insight into human nervous tissues is needed to generate next-generation therapeutics for neurological disorders such as chronic pain. We obtained human dorsal root ganglia (hDRG) samples from organ donors and performed RNA-sequencing (RNA-seq) to study the hDRG transcriptional landscape, systematically comparing it with publicly available data from a variety of human and orthologous mouse tissues, including mouse DRG (mDRG). We characterized the hDRG transcriptional profile in terms of tissue-restricted gene coexpression patterns and putative transcriptional regulators, and formulated an information-theoretic framework to quantify DRG enrichment. Relevant gene families and pathways were also analyzed, including transcription factors, G-protein-coupled receptors, and ion channels. Our analyses reveal an hDRG-enriched protein-coding gene set (∼140), some of which have not been described in the context of DRG or pain signaling. Most of these show conserved enrichment in mDRG and were mined for known drug-gene product interactions. Conserved enrichment of the vast majority of transcription factors suggests that the mDRG is a faithful model system for studying hDRG, because of evolutionarily conserved regulatory programs. Comparison of hDRG and tibial nerve transcriptomes suggests trafficking of neuronal mRNA to axons in adult hDRG, and are consistent with studies of axonal transport in rodent sensory neurons. We present our work as an online, searchable repository (https://www.utdallas.edu/bbs/painneurosciencelab/sensoryomics/drgtxome), creating a valuable resource for the community. Our analyses provide insight into DRG biology for guiding development of novel therapeutics and a blueprint for cross-species transcriptomic analyses.
Conflict of interest statement
Figures





References
-
- Abaffy T. Human Olfactory Receptors Expression and Their Role in Non-Olfactory Tissues-A Mini-Review. Journal of Pharmacogenomics & Pharmacoproteomics. 2015;6(4):1.
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25(1):25–29. - PMC - PubMed
-
- Atluri S, Sudarshan G, Manchikanti L. Assessment of the trends in medical use and misuse of opioid analgesics from 2004 to 2011. Pain Physician. 2014;17(2):E119–128. - PubMed
-
- Bannwarth B, Kostine M. Targeting nerve growth factor (NGF) for pain management: what does the future hold for NGF antagonists? Drugs. 2014;74(6):619–626. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

