High-Throughput Sequencing Analysis of the Actinobacterial Spatial Diversity in Moonmilk Deposits
- PMID: 29561792
- PMCID: PMC6023079
- DOI: 10.3390/antibiotics7020027
High-Throughput Sequencing Analysis of the Actinobacterial Spatial Diversity in Moonmilk Deposits
Abstract
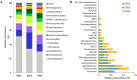
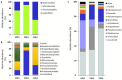
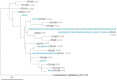
Moonmilk are cave carbonate deposits that host a rich microbiome, including antibiotic-producing Actinobacteria, making these speleothems appealing for bioprospecting. Here, we investigated the taxonomic profile of the actinobacterial community of three moonmilk deposits of the cave "Grotte des Collemboles" via high-throughput sequencing of 16S rRNA amplicons. Actinobacteria was the most common phylum after Proteobacteria, ranging from 9% to 23% of the total bacterial population. Next to actinobacterial operational taxonomic units (OTUs) attributed to uncultured organisms at the genus level (~44%), we identified 47 actinobacterial genera with Rhodoccocus (4 OTUs, 17%) and Pseudonocardia (9 OTUs, ~16%) as the most abundant in terms of the absolute number of sequences. Streptomycetes presented the highest diversity (19 OTUs, 3%), with most of the OTUs unlinked to the culturable Streptomyces strains that were previously isolated from the same deposits. Furthermore, 43% of the OTUs were shared between the three studied collection points, while 34% were exclusive to one deposit, indicating that distinct speleothems host their own population, despite their nearby localization. This important spatial diversity suggests that prospecting within different moonmilk deposits should result in the isolation of unique and novel Actinobacteria. These speleothems also host a wide range of non-streptomycetes antibiotic-producing genera, and should therefore be subjected to methodologies for isolating rare Actinobacteria.
Keywords: Actinobacteria; Illumina sequencing; Streptomyces; antibiotics; geomicrobiology; microbiome diversity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits.Antibiotics (Basel). 2018 Mar 22;7(2):28. doi: 10.3390/antibiotics7020028. Antibiotics (Basel). 2018. PMID: 29565274 Free PMC article.
-
A Phenotypic and Genotypic Analysis of the Antimicrobial Potential of Cultivable Streptomyces Isolated from Cave Moonmilk Deposits.Front Microbiol. 2016 Sep 21;7:1455. doi: 10.3389/fmicb.2016.01455. eCollection 2016. Front Microbiol. 2016. PMID: 27708627 Free PMC article.
-
Diversity of Bacteria and the Characteristics of Actinobacteria Community Structure in Badain Jaran Desert and Tengger Desert of China.Front Microbiol. 2018 May 23;9:1068. doi: 10.3389/fmicb.2018.01068. eCollection 2018. Front Microbiol. 2018. PMID: 29875762 Free PMC article.
-
Korean indigenous bacterial species with valid names belonging to the phylum Actinobacteria.J Microbiol. 2016 Dec;54(12):789-795. doi: 10.1007/s12275-016-6446-4. Epub 2016 Nov 26. J Microbiol. 2016. PMID: 27888457 Review.
-
Cave Actinobacteria as Producers of Bioactive Metabolites.Front Microbiol. 2019 Mar 22;10:387. doi: 10.3389/fmicb.2019.00387. eCollection 2019. Front Microbiol. 2019. PMID: 30967844 Free PMC article. Review.
Cited by
-
Calcite moonmilk of microbial origin in the Etruscan Tomba degli Scudi in Tarquinia, Italy.Sci Rep. 2018 Oct 26;8(1):15839. doi: 10.1038/s41598-018-34134-y. Sci Rep. 2018. PMID: 30367083 Free PMC article.
-
Structure of New Ferroverdins Recruiting Unconventional Ferrous Iron Chelating Agents.Biomolecules. 2022 May 26;12(6):752. doi: 10.3390/biom12060752. Biomolecules. 2022. PMID: 35740878 Free PMC article.
-
Whole Genome Sequencing and Metabolomic Study of Cave Streptomyces Isolates ICC1 and ICC4.Front Microbiol. 2019 May 10;10:1020. doi: 10.3389/fmicb.2019.01020. eCollection 2019. Front Microbiol. 2019. PMID: 31134037 Free PMC article.
-
Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits.Antibiotics (Basel). 2018 Mar 22;7(2):28. doi: 10.3390/antibiotics7020028. Antibiotics (Basel). 2018. PMID: 29565274 Free PMC article.
-
Shotgun metagenomic sequencing from Manao-Pee cave, Thailand, reveals insight into the microbial community structure and its metabolic potential.BMC Microbiol. 2019 Jun 27;19(1):144. doi: 10.1186/s12866-019-1521-8. BMC Microbiol. 2019. PMID: 31248378 Free PMC article.
References
-
- Engel A.S. Geomicrobiology: Molecular and Environmental Perspective. Springer; Berlin, Germany: 2010. Microbial diversity of cave ecosystems.
-
- Barton H.A., Northup D.E. Geomicrobiology in cave environments: Past, current and future perspectives. J. Cave Karst Stud. 2007;69:163–178.
-
- Jurado V., Kroppenstedt R.M., Saiz-Jimenez C., Klenk H.P., Mouniée D., Laiz L., Couble A., Pötter G., Boiron P., Rodríguez-Nava V. Hoyosella altamirensis gen. nov., sp. nov., a new member of the order Actinomycetales isolated from a cave biofilm. Int. J. Syst. Evol. Microbiol. 2009;59:3105–3110. doi: 10.1099/ijs.0.008664-0. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

