KIBRA (WWC1) Is a Metastasis Suppressor Gene Affected by Chromosome 5q Loss in Triple-Negative Breast Cancer
- PMID: 29562176
- PMCID: PMC5873529
- DOI: 10.1016/j.celrep.2018.02.095
KIBRA (WWC1) Is a Metastasis Suppressor Gene Affected by Chromosome 5q Loss in Triple-Negative Breast Cancer
Abstract
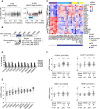
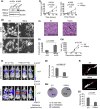
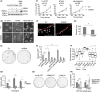
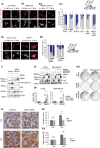
Triple-negative breast cancers (TNBCs) display a complex spectrum of mutations and chromosomal aberrations. Chromosome 5q (5q) loss is detected in up to 70% of TNBCs, but little is known regarding the genetic drivers associated with this event. Here, we show somatic deletion of a region syntenic with human 5q33.2-35.3 in a mouse model of TNBC. Mechanistically, we identify KIBRA as a major factor contributing to the effects of 5q loss on tumor growth and metastatic progression. Re-expression of KIBRA impairs metastasis in vivo and inhibits tumorsphere formation by TNBC cells in vitro. KIBRA functions co-operatively with the protein tyrosine phosphatase PTPN14 to trigger mechanotransduction-regulated signals that inhibit the nuclear localization of oncogenic transcriptional co-activators YAP/TAZ. Our results argue that the selective advantage produced by 5q loss involves reduced dosage of KIBRA, promoting oncogenic functioning of YAP/TAZ in TNBC.
Keywords: KIBRA; PTPN14; RHOA signaling; WWC1; YAP/TAZ; chr5q; mechanotransduction; metastasis; triple-negative breast cancer; tumorspheres.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. - PubMed
-
- Bartucci M., Dattilo R., Moriconi C., Pagliuca A., Mottolese M., Federici G., Benedetto A.D., Todaro M., Stassi G., Sperati F. TAZ is required for metastatic activity and chemoresistance of breast cancer stem cells. Oncogene. 2015;34:681–690. - PubMed
-
- Baumgartner R., Poernbacher I., Buser N., Hafen E., Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell. 2010;18:309–316. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

