TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis
- PMID: 29562314
- PMCID: PMC5917749
- DOI: 10.1093/brain/awy062
TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis
Abstract
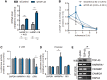
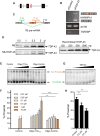
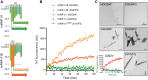
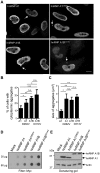
See Fratta and Isaacs (doi:10.1093/brain/awy091) for a scientific commentary on this article.The RNA binding proteins TDP-43 (encoded by TARDBP) and hnRNP A1 (HNRNPA1) are each mutated in certain amyotrophic lateral sclerosis cases and are often mislocalized in cytoplasmic aggregates within motor neurons of affected patients. Cytoplasmic inclusions of TDP-43, which are accompanied by a depletion of nuclear TDP-43, are observed in most amyotrophic lateral sclerosis cases and nearly half of frontotemporal dementia cases. Here, we report that TDP-43 binds HNRNPA1 pre-mRNA and modulates its splicing, and that depletion of nuclear TDP-43 results in increased inclusion of a cassette exon in the HNRNPA1 transcript, and consequently elevated protein levels of an isoform containing an elongated prion-like domain, referred to as hnRNP A1B. Combined in vivo and in vitro approaches demonstrated greater fibrillization propensity for hnRNP A1B, which drives protein aggregation and is toxic to cells. Moreover, amyotrophic lateral sclerosis patients with documented TDP-43 pathology showed neuronal hnRNP A1B cytoplasmic accumulation, indicating that TDP-43 mislocalization may contribute to neuronal vulnerability and loss via altered HNRNPA1 pre-mRNA splicing and function. Given that TDP-43 and hnRNP A1 each bind, and thus modulate, a third of the transcriptome, our data suggest a much broader disruption in RNA metabolism than previously considered.
Figures


Comment in
-
The snowball effect of RNA binding protein dysfunction in amyotrophic lateral sclerosis.Brain. 2018 May 1;141(5):1236-1238. doi: 10.1093/brain/awy091. Brain. 2018. PMID: 29701791 Free PMC article.
-
TDP-43 mutations increase HNRNP A1-7B through gain of splicing function.Brain. 2018 Dec 1;141(12):e83. doi: 10.1093/brain/awy260. Brain. 2018. PMID: 30364928 No abstract available.
-
Reply: TDP-43 mutations increase HNRNP A1-7B through gain of splicing function.Brain. 2018 Dec 1;141(12):e84. doi: 10.1093/brain/awy261. Brain. 2018. PMID: 30364932 Free PMC article. No abstract available.
Similar articles
-
Loss of hnRNPA1 in ALS spinal cord motor neurons with TDP-43-positive inclusions.Neuropathology. 2015 Feb;35(1):37-43. doi: 10.1111/neup.12153. Epub 2014 Oct 22. Neuropathology. 2015. PMID: 25338872
-
Increased cytoplasmic TARDBP mRNA in affected spinal motor neurons in ALS caused by abnormal autoregulation of TDP-43.Nucleic Acids Res. 2016 Jul 8;44(12):5820-36. doi: 10.1093/nar/gkw499. Epub 2016 Jun 2. Nucleic Acids Res. 2016. PMID: 27257061 Free PMC article.
-
Quantification of the Relative Contributions of Loss-of-function and Gain-of-function Mechanisms in TAR DNA-binding Protein 43 (TDP-43) Proteinopathies.J Biol Chem. 2016 Sep 9;291(37):19437-48. doi: 10.1074/jbc.M116.737726. Epub 2016 Jul 21. J Biol Chem. 2016. PMID: 27445339 Free PMC article.
-
The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients.Brain Res. 2014 Oct 10;1584:28-38. doi: 10.1016/j.brainres.2013.12.011. Epub 2013 Dec 16. Brain Res. 2014. PMID: 24355598 Review.
-
The debated toxic role of aggregated TDP-43 in amyotrophic lateral sclerosis: a resolution in sight?Brain. 2019 May 1;142(5):1176-1194. doi: 10.1093/brain/awz078. Brain. 2019. PMID: 30938443 Free PMC article. Review.
Cited by
-
TDP-43 and other hnRNPs regulate cryptic exon inclusion of a key ALS/FTD risk gene, UNC13A.PLoS Biol. 2023 Mar 17;21(3):e3002028. doi: 10.1371/journal.pbio.3002028. eCollection 2023 Mar. PLoS Biol. 2023. PMID: 36930682 Free PMC article.
-
hnRNP R negatively regulates transcription by modulating the association of P-TEFb with 7SK and BRD4.EMBO Rep. 2022 Sep 5;23(9):e55432. doi: 10.15252/embr.202255432. Epub 2022 Jul 20. EMBO Rep. 2022. PMID: 35856391 Free PMC article.
-
The effects of N6-methyladenosine RNA methylation on the nervous system.Mol Cell Biochem. 2023 Dec;478(12):2657-2669. doi: 10.1007/s11010-023-04691-6. Epub 2023 Mar 10. Mol Cell Biochem. 2023. PMID: 36899139 Review.
-
Targeted-Deletion of a Tiny Sequence via Prime Editing to Restore SMN Expression.Int J Mol Sci. 2022 Jul 19;23(14):7941. doi: 10.3390/ijms23147941. Int J Mol Sci. 2022. PMID: 35887289 Free PMC article.
-
Could an Impairment in Local Translation of mRNAs in Glia be Contributing to Pathogenesis in ALS?Front Mol Neurosci. 2019 May 21;12:124. doi: 10.3389/fnmol.2019.00124. eCollection 2019. Front Mol Neurosci. 2019. PMID: 31164803 Free PMC article. Review.
References
-
- Bekenstein U, Soreq H. Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci 2013; 56: 436–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

