Structural basis for LeishIF4E-1 modulation by an interacting protein in the human parasite Leishmania major
- PMID: 29562352
- PMCID: PMC5909430
- DOI: 10.1093/nar/gky194
Structural basis for LeishIF4E-1 modulation by an interacting protein in the human parasite Leishmania major
Abstract
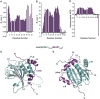
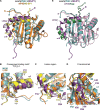
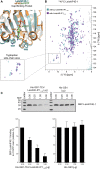
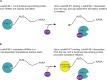
Leishmania parasites are unicellular pathogens that are transmitted to humans through the bite of infected sandflies. Most of the regulation of their gene expression occurs post-transcriptionally, and the different patterns of gene expression required throughout the parasites' life cycle are regulated at the level of translation. Here, we report the X-ray crystal structure of the Leishmania cap-binding isoform 1, LeishIF4E-1, bound to a protein fragment of previously unknown function, Leish4E-IP1, that binds tightly to LeishIF4E-1. The molecular structure, coupled to NMR spectroscopy experiments and in vitro cap-binding assays, reveal that Leish4E-IP1 allosterically destabilizes the binding of LeishIF4E-1 to the 5' mRNA cap. We propose mechanisms through which Leish4E-IP1-mediated LeishIF4E-1 inhibition could regulate translation initiation in the human parasite.
Figures
Similar articles
-
LeishIF4E-5 Is a Promastigote-Specific Cap-Binding Protein in Leishmania.Int J Mol Sci. 2021 Apr 12;22(8):3979. doi: 10.3390/ijms22083979. Int J Mol Sci. 2021. PMID: 33921489 Free PMC article.
-
Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania.Eukaryot Cell. 2006 Dec;5(12):1969-79. doi: 10.1128/EC.00230-06. Epub 2006 Oct 13. Eukaryot Cell. 2006. PMID: 17041189 Free PMC article.
-
A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways.Nucleic Acids Res. 2011 Oct;39(19):8404-15. doi: 10.1093/nar/gkr555. Epub 2011 Jul 14. Nucleic Acids Res. 2011. PMID: 21764780 Free PMC article.
-
Structure and functions of the translation initiation factor eIF4E and its role in cancer development and treatment.J Genet Genomics. 2018 Jan 20;45(1):13-24. doi: 10.1016/j.jgg.2018.01.003. Epub 2018 Feb 1. J Genet Genomics. 2018. PMID: 29396141 Review.
-
Translational control of mRNAs by 3'-Untranslated region binding proteins.BMB Rep. 2017 Apr;50(4):194-200. doi: 10.5483/bmbrep.2017.50.4.040. BMB Rep. 2017. PMID: 28287067 Free PMC article. Review.
Cited by
-
eIF4E and Interactors from Unicellular Eukaryotes.Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
-
LeishIF4E-5 Is a Promastigote-Specific Cap-Binding Protein in Leishmania.Int J Mol Sci. 2021 Apr 12;22(8):3979. doi: 10.3390/ijms22083979. Int J Mol Sci. 2021. PMID: 33921489 Free PMC article.
-
Regulation of gene expression in trypanosomatids: living with polycistronic transcription.Open Biol. 2019 Jun 28;9(6):190072. doi: 10.1098/rsob.190072. Epub 2019 Jun 5. Open Biol. 2019. PMID: 31164043 Free PMC article.
-
The EIF4E1-4EIP cap-binding complex of Trypanosoma brucei interacts with the terminal uridylyl transferase TUT3.PLoS One. 2021 Nov 22;16(11):e0258903. doi: 10.1371/journal.pone.0258903. eCollection 2021. PLoS One. 2021. PMID: 34807934 Free PMC article.
-
Taking a re-look at cap-binding signatures of the mRNA cap-binding protein eIF4E orthologues in trypanosomatids.Mol Cell Biochem. 2021 Feb;476(2):1037-1049. doi: 10.1007/s11010-020-03970-w. Epub 2020 Nov 10. Mol Cell Biochem. 2021. PMID: 33169189 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

