Agnoprotein Is an Essential Egress Factor during BK Polyomavirus Infection
- PMID: 29562663
- PMCID: PMC5877763
- DOI: 10.3390/ijms19030902
Agnoprotein Is an Essential Egress Factor during BK Polyomavirus Infection
Abstract
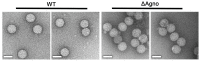
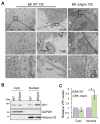
BK polyomavirus (BKPyV; hereafter referred to as BK) causes a lifelong chronic infection and is associated with debilitating disease in kidney transplant recipients. Despite its importance, aspects of the virus life cycle remain poorly understood. In addition to the structural proteins, the late region of the BK genome encodes for an auxiliary protein called agnoprotein. Studies on other polyomavirus agnoproteins have suggested that the protein may contribute to virion infectivity. Here, we demonstrate an essential role for agnoprotein in BK virus release. Viruses lacking agnoprotein fail to release from host cells and do not propagate to wild-type levels. Despite this, agnoprotein is not essential for virion infectivity or morphogenesis. Instead, agnoprotein expression correlates with nuclear egress of BK virions. We demonstrate that the agnoprotein binding partner α-soluble N-ethylmaleimide sensitive fusion (NSF) attachment protein (α-SNAP) is necessary for BK virion release, and siRNA knockdown of α-SNAP prevents nuclear release of wild-type BK virions. These data highlight a novel role for agnoprotein and begin to reveal the mechanism by which polyomaviruses leave an infected cell.
Keywords: agnoprotein; polyomavirus; virus exit.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

