Transcriptomic analysis of Perilla frutescens seed to insight into the biosynthesis and metabolic of unsaturated fatty acids
- PMID: 29562889
- PMCID: PMC5863459
- DOI: 10.1186/s12864-018-4595-z
Transcriptomic analysis of Perilla frutescens seed to insight into the biosynthesis and metabolic of unsaturated fatty acids
Abstract
Background: Perilla frutescens is well known for its high α-linolenic acid (ALA) accumulation in seeds and medicinal values as well as a source of edible and general-purpose oils. However, the regulatory mechanisms of the biosynthesis of fatty acid in its seeds remain poorly understood due to the lacking of sequenced genome. For better understanding the regulation of lipid metabolism and further increase its oil content or modify oil composition, time-course transcriptome and lipid composition analyses were performed.
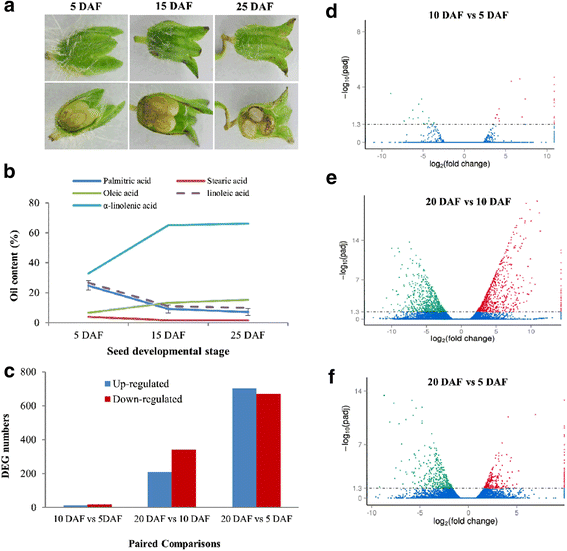
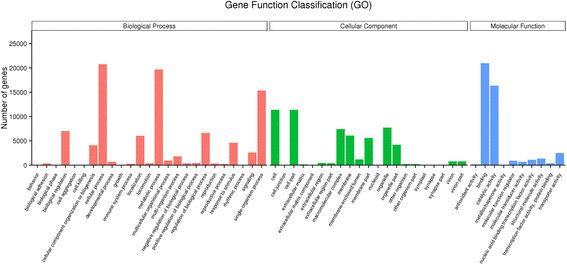
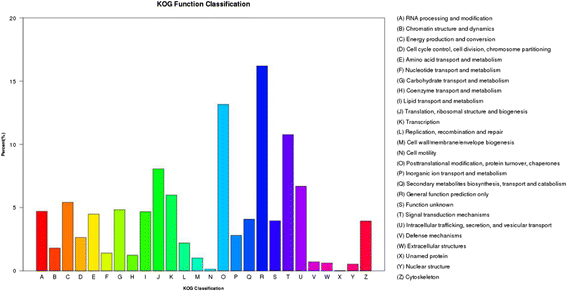
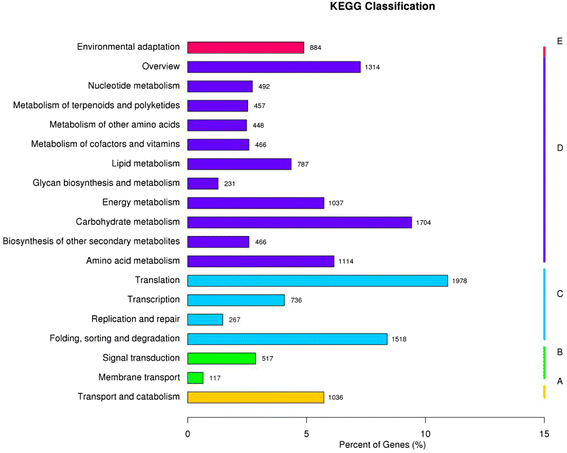
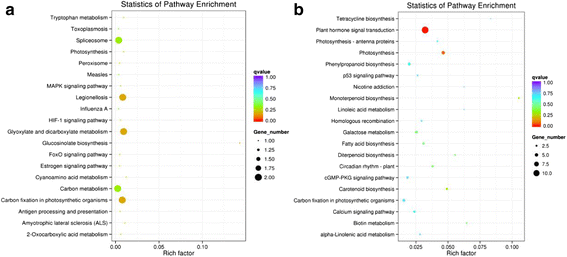
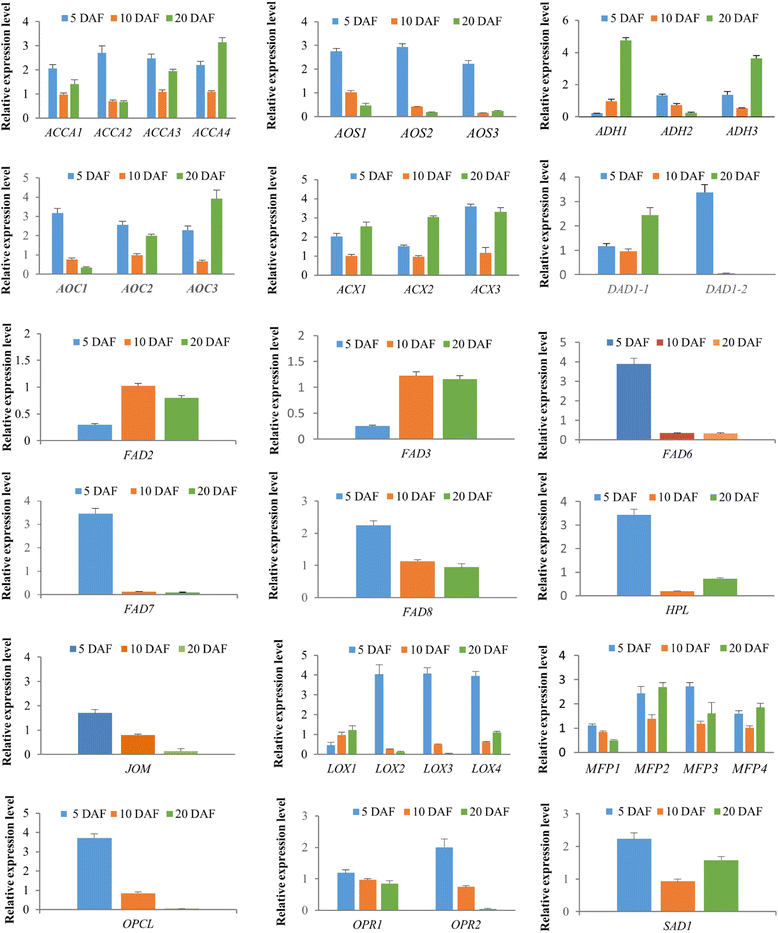
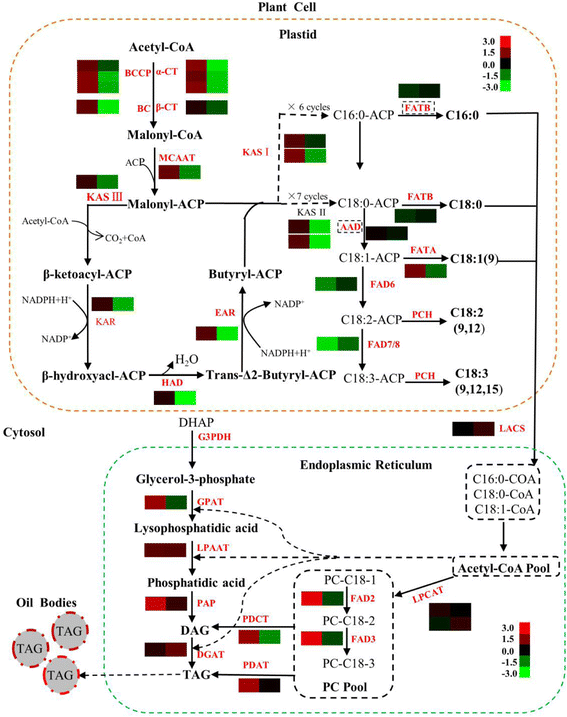
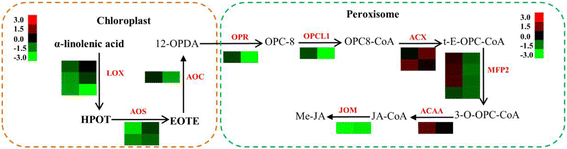
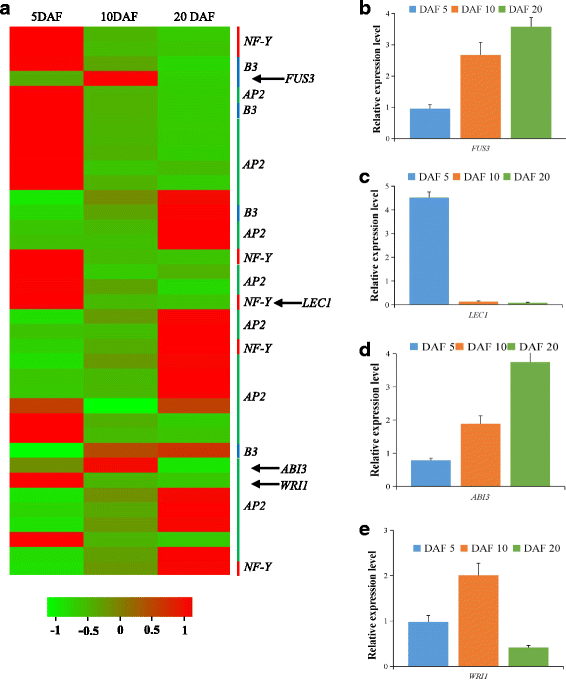
Results: Analysis of fatty acid content and composition showed that the α-linolenic acid and oleic acid accumulated rapidly from 5 DAF to 15 DAF and then kept relatively stable. However, the amount of palmitic acid and linoleic acid decreased quickly from 5 DAF to 15DAF. No significant variation of stearic acid content was observed from 5 DAF to 25DAF. Our transcriptome data analyses revealed that 110,176 unigenes were generated from six seed libraries at 5, 10, 20 DAF. Of these, 53 (31 up, 22 down) and 653 (259 up, 394 down) genes showed temporal and differentially expression during the seed development in 5 DAF vs 10 DAF, 20 vs 10 DAF, respectively. The differentially expressed genes were annotated and found to be involved in distinct functional categories and metabolic pathways. Deep mining of transcriptome data led to the identification of key genes involved in fatty acid and triacylglycerol biosynthesis and metabolism. Thirty seven members of transcription factor family AP2, B3 and NFYB putatively involved in oil synthesis and deposition were differentially expressed during seed development. The results of qRT-PCR for selected genes showed a strong positive correlation with the expression abundance measured in RNA-seq analysis.
Conclusions: The present study provides valuable genomic resources for characterizing Perilla seed gene expression at the transcriptional level and will extend our understanding of the complex molecular and cellular events of oil biosynthesis and accumulation in oilseed crops.
Keywords: Fatty acid biosynthesis; Gene expression profiling; Perilla frutescens; RNA-seq; Seed development.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Park YJ, Dixit A, Ma KH, Lee JK, Lee MH, Chung CS, Nitta M, Okuno K, Kim TS, Cho EG. Evaluation of genetic diversity and relationships within an on-farm collection of Perilla frutescens (L.) Britt. Using microsatellite markers. Genet Resour Crop Evol. 2008;55(4):523–535. doi: 10.1007/s10722-007-9258-x. - DOI
-
- Shin HS, Kim SW. Lipid composition of perilla seed. J Am Oil Chem Soc. 1994;71(6):619–622. doi: 10.1007/BF02540589. - DOI
-
- Ciftci ON, Przybylski R, Rudzińska M. Lipid components of flax, perilla, and chia seeds. Eur J Lipid Sci Technol. 2012;114(7):794–800. doi: 10.1002/ejlt.201100207. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

