Balance between MAT2A intron detention and splicing is determined cotranscriptionally
- PMID: 29563249
- PMCID: PMC5959247
- DOI: 10.1261/rna.064899.117
Balance between MAT2A intron detention and splicing is determined cotranscriptionally
Abstract
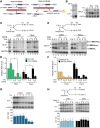
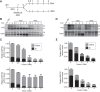
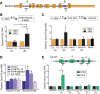
Transcriptome analysis of human cells has revealed that intron retention controls the expression of a large number of genes with diverse cellular functions. Detained introns (DI) constitute a subgroup of transcripts with retained introns that are not exported to the cytoplasm but instead remain in the nucleus. Previous studies reported that the splicing of DIs in the CLK1 transcript is post-transcriptionally induced to produce mature mRNA in the absence of new transcription. Thus, CLK1-DI serves as a precursor or "reservoir" for the CLK1 mRNA. However, whether this is a universal mechanism for gene regulation by intron detention remains unknown. The MAT2A gene encodes S-adenosylmethionine (SAM) synthetase and it contains a DI that is regulated in response to intracellular SAM levels. We used three independent assays to assess the precursor-product relationship between MAT2A-DI and MAT2A mRNA. In contrast to CLK1-DI, these data support a model in which the MAT2A-DI transcript is not a precursor to mRNA but is instead a "dead-end" RNA fated for nuclear decay. Additionally, we show that in SAM-deprived conditions the cotranscriptional splicing of MAT2A detained introns increases. We conclude that polyadenylated RNAs with DIs can have at least two distinct fates. They can serve as nuclear reservoirs of pre-mRNAs available for rapid induction by the cell, or they constitute dead-end RNAs that are degraded in the nucleus.
Keywords: MAT2A; S-adenosylmethionine; intron detention; intron retention; splicing.
© 2018 Pendleton et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

References
-
- Braun CJ, Stanciu M, Boutz PL, Patterson JC, Calligaris D, Higuchi F, Neupane R, Fenoglio S, Cahill DP, Wakimoto H, et al. 2017. Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell 32: 411–426.e11. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
