Dynamical origins of heat capacity changes in enzyme-catalysed reactions
- PMID: 29563521
- PMCID: PMC5862990
- DOI: 10.1038/s41467-018-03597-y
Dynamical origins of heat capacity changes in enzyme-catalysed reactions
Abstract
Heat capacity changes are emerging as essential for explaining the temperature dependence of enzyme-catalysed reaction rates. This has important implications for enzyme kinetics, thermoadaptation and evolution, but the physical basis of these heat capacity changes is unknown. Here we show by a combination of experiment and simulation, for two quite distinct enzymes (dimeric ketosteroid isomerase and monomeric alpha-glucosidase), that the activation heat capacity change for the catalysed reaction can be predicted through atomistic molecular dynamics simulations. The simulations reveal subtle and surprising underlying dynamical changes: tightening of loops around the active site is observed, along with changes in energetic fluctuations across the whole enzyme including important contributions from oligomeric neighbours and domains distal to the active site. This has general implications for understanding enzyme catalysis and demonstrating a direct connection between functionally important microscopic dynamics and macroscopically measurable quantities.
Conflict of interest statement
The authors declare no competing interests.
Figures
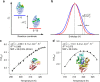
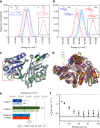
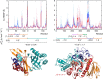
References
-
- Arrhenius S. Über die Reaktionsgeschwindigkeit bei der Inversion von Rohrzucker durch Säuren. Z. Phys. Chem. 1889;4:226–248.
-
- Eyring H. The activated complex in chemical reactions. J. Chem. Phys. 1935;3:107–115. doi: 10.1063/1.1749604. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

