Mutation patterns identify adult patients with de novo acute myeloid leukemia aged 60 years or older who respond favorably to standard chemotherapy: an analysis of Alliance studies
- PMID: 29563537
- PMCID: PMC5992022
- DOI: 10.1038/s41375-018-0068-2
Mutation patterns identify adult patients with de novo acute myeloid leukemia aged 60 years or older who respond favorably to standard chemotherapy: an analysis of Alliance studies
Abstract
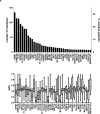
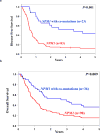
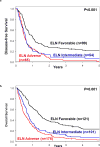
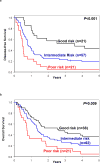
Thus far, only 5-15% of AML patients aged ≥60 years are cured with chemotherapy. Identification of patients who are less (more) likely to respond to standard chemotherapy might enable early risk stratification toward alternative treatment regimens. We used a next-generation sequencing panel of 80 cancer- and/or leukemia-associated genes to profile molecularly 423 older patients with de novo AML. Using variables identified in multivariable models and co-occurring mutations in NPM1-mutated AML, we classified the patients into good-, intermediate-, and poor-risk groups for complete remission (CR) attainment, disease-free (DFS), and overall survival (OS). Whereas 81% of good-risk patients (comprising NPM1-mutated patients harboring mutations in chromatin remodeling, cohesin complex, methylation-related, spliceosome, and/or RAS pathway genes, FLT3-TKD, and/or patients without FLT3-ITD) achieved a CR, only 32% of poor-risk patients (with U2AF1, WT1 mutations and/or complex karyotype) did. Intermediate-risk patients had a 50% CR rate. Similarly, using NPM1 co-mutation patterns and SF1 mutation status, we identified patients with favorable DFS and OS 3-year rates of 46% and 45%, respectively. Patients with adverse genetic features had DFS and OS rates of only 2% and 4%. We show that application of our proposed criteria may refine the 2017 European LeukemiaNet classification for older patients treated with chemotherapy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. - PubMed
-
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U10 CA101140/CA/NCI NIH HHS/United States
- R35 CA197734/CA/NCI NIH HHS/United States
- K05 CA131504/CA/NCI NIH HHS/United States
- U10 CA180882/CA/NCI NIH HHS/United States
- U10 CA031946/CA/NCI NIH HHS/United States
- U10 CA180850/CA/NCI NIH HHS/United States
- U10 CA180821/CA/NCI NIH HHS/United States
- P50 CA140158/CA/NCI NIH HHS/United States
- UG1 CA189850/CA/NCI NIH HHS/United States
- U10 CA180867/CA/NCI NIH HHS/United States
- U24 CA196171/CA/NCI NIH HHS/United States
- U10 CA180866/CA/NCI NIH HHS/United States
- U10 CA003927/CA/NCI NIH HHS/United States
- R01 CA152045/CA/NCI NIH HHS/United States
- U10 CA180861/CA/NCI NIH HHS/United States
- U10 CA041287/CA/NCI NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

