Cervical microbiome is altered in cervical intraepithelial neoplasia after loop electrosurgical excision procedure in china
- PMID: 29563617
- PMCID: PMC5862885
- DOI: 10.1038/s41598-018-23389-0
Cervical microbiome is altered in cervical intraepithelial neoplasia after loop electrosurgical excision procedure in china
Abstract
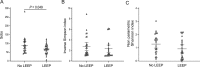
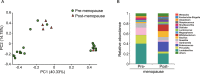
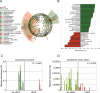
Although human papillomavirus (HPV) infection is a major cause leading to the development of cervical intraepithelial neoplasia (CIN), the relationship between genital microbiome and HPV persistence/clearance is not well established. Loop electrosurgical excision procedure (LEEP) is one of standard treatments of CIN 2/3 globally, yet little is known about how the LEEP influence genital microbiota. We conducted a prospective study of 26 patients with CIN2/3 who underwent analysis of cervical microbiome before and after 3 months of LEEP treatment. Cervical swabs were collected, and microbiomes were analyzed by 16S ribosomal RNA gene sequencing. A decrease of cervical microbial diversity was observed after 3 months of LEEP treatment. Notably, a significant shift from community type of a Prevotella-containing and lack of a consistent dominant species to lactobacillus iners dominated microbiome correlated with LEEP. Particularly, Leptotrichia and clostridium were further decreased after LEEP treatment (P = 0.049 and P = 0.002, respectively). Our results suggest that the cervical microbiome is altered after LEEP treatment in patients with CIN2/3. Further studies with larger sample sizes are needed to validate these findings.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Khan MJ, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. Journal of the National Cancer Institute. 2005;97:1072–1079. doi: 10.1093/jnci/dji187. - DOI - PubMed
-
- Oh HY, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2015;21(674):e671–679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

