Molecular Basis of Encapsidation of Hepatitis C Virus Genome
- PMID: 29563905
- PMCID: PMC5845887
- DOI: 10.3389/fmicb.2018.00396
Molecular Basis of Encapsidation of Hepatitis C Virus Genome
Abstract
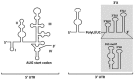
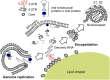
Hepatitis C virus (HCV), a major etiologic agent of human liver diseases, is a positive-sense single-stranded RNA virus and is classified in the Flaviviridae family. Although research findings for the assembly of HCV particles are accumulating due to development of HCV cell culture system, the mechanism(s) by which the HCV genome becomes encapsidated remains largely unclear. In general, viral RNA represents only a small fraction of the RNA molecules in the cells infected with RNA viruses, but the viral genomic RNA is considered to selectively packaged into virions. It was recently demonstrated that HCV RNAs containing 3' end of the genome are selectively incorporated into virus particles during the assembly process and the 3' untranslated region functions as a cis-acting element for RNA packaging. Here, we discuss the molecular basis of RNA encapsidation of HCV and classical flaviviruses, contrast with the packaging mechanism of HIV-1.
Keywords: cis-acting element; encapsidation; hepatitis C virus; packaging signal; virion assembly.
Figures
Similar articles
-
Involvement of the 3' Untranslated Region in Encapsidation of the Hepatitis C Virus.PLoS Pathog. 2016 Feb 11;12(2):e1005441. doi: 10.1371/journal.ppat.1005441. eCollection 2016 Feb. PLoS Pathog. 2016. PMID: 26867128 Free PMC article.
-
Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles.J Virol. 2008 Jul;82(14):7034-46. doi: 10.1128/JVI.00118-08. Epub 2008 May 14. J Virol. 2008. PMID: 18480457 Free PMC article.
-
A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation.Cell Host Microbe. 2013 Feb 13;13(2):181-92. doi: 10.1016/j.chom.2013.01.007. Cell Host Microbe. 2013. PMID: 23414758 Free PMC article.
-
Signals Involved in Regulation of Hepatitis C Virus RNA Genome Translation and Replication.Front Microbiol. 2018 Mar 12;9:395. doi: 10.3389/fmicb.2018.00395. eCollection 2018. Front Microbiol. 2018. PMID: 29593672 Free PMC article. Review.
-
cis-Acting RNA elements in the hepatitis C virus RNA genome.Virus Res. 2015 Aug 3;206:90-8. doi: 10.1016/j.virusres.2014.12.029. Epub 2015 Jan 7. Virus Res. 2015. PMID: 25576644 Free PMC article. Review.
Cited by
-
Structural phylogenetic analysis reveals lineage-specific RNA repetitive structural motifs in all coronaviruses and associated variations in SARS-CoV-2.Virus Evol. 2021 Jun 16;7(1):veab021. doi: 10.1093/ve/veab021. eCollection 2021 Jan. Virus Evol. 2021. PMID: 34141447 Free PMC article.
-
Pathogens and Carcinogenesis: A Review.Biology (Basel). 2021 Jun 15;10(6):533. doi: 10.3390/biology10060533. Biology (Basel). 2021. PMID: 34203649 Free PMC article. Review.
-
Impact of hepatitis C virus genotype on the efficacy of the direct-acting antivirals in chronic kidney disease patients in West Bengal, India.BMC Infect Dis. 2025 May 16;25(1):706. doi: 10.1186/s12879-025-10947-x. BMC Infect Dis. 2025. PMID: 40380327 Free PMC article.
-
Immune Microenvironment and Immunotherapeutic Management in Virus-Associated Digestive System Tumors.Int J Mol Sci. 2022 Nov 6;23(21):13612. doi: 10.3390/ijms232113612. Int J Mol Sci. 2022. PMID: 36362398 Free PMC article. Review.
-
Nup98 Is Subverted from Annulate Lamellae by Hepatitis C Virus Core Protein to Foster Viral Assembly.mBio. 2022 Apr 26;13(2):e0292321. doi: 10.1128/mbio.02923-21. Epub 2022 Mar 8. mBio. 2022. PMID: 35258330 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

