An LaeA- and BrlA-Dependent Cellular Network Governs Tissue-Specific Secondary Metabolism in the Human Pathogen Aspergillus fumigatus
- PMID: 29564395
- PMCID: PMC5853485
- DOI: 10.1128/mSphere.00050-18
An LaeA- and BrlA-Dependent Cellular Network Governs Tissue-Specific Secondary Metabolism in the Human Pathogen Aspergillus fumigatus
Abstract
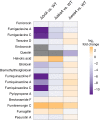
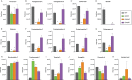
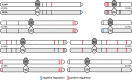
Biosynthesis of many ecologically important secondary metabolites (SMs) in filamentous fungi is controlled by several global transcriptional regulators, like the chromatin modifier LaeA, and tied to both development and vegetative growth. In Aspergillus molds, asexual development is regulated by the BrlA > AbaA > WetA transcriptional cascade. To elucidate BrlA pathway involvement in SM regulation, we examined the transcriptional and metabolic profiles of ΔbrlA, ΔabaA, and ΔwetA mutant and wild-type strains of the human pathogen Aspergillus fumigatus. We find that BrlA, in addition to regulating production of developmental SMs, regulates vegetative SMs and the SrbA-regulated hypoxia stress response in a concordant fashion to LaeA. We further show that the transcriptional and metabolic equivalence of the ΔbrlA and ΔlaeA mutations is mediated by an LaeA requirement preventing heterochromatic marks in the brlA promoter. These results provide a framework for the cellular network regulating not only fungal SMs but diverse cellular processes linked to virulence of this pathogen. IMPORTANCE Filamentous fungi produce a spectacular variety of small molecules, commonly known as secondary or specialized metabolites (SMs), which are critical to their ecologies and lifestyles (e.g., penicillin, cyclosporine, and aflatoxin). Elucidation of the regulatory network that governs SM production is a major question of both fundamental and applied research relevance. To shed light on the relationship between regulation of development and regulation of secondary metabolism in filamentous fungi, we performed global transcriptomic and metabolomic analyses on mutant and wild-type strains of the human pathogen Aspergillus fumigatus under conditions previously shown to induce the production of both vegetative growth-specific and asexual development-specific SMs. We find that the gene brlA, previously known as a master regulator of asexual development, is also a master regulator of secondary metabolism and other cellular processes. We further show that brlA regulation of SM is mediated by laeA, one of the master regulators of SM, providing a framework for the cellular network regulating not only fungal SMs but diverse cellular processes linked to virulence of this pathogen.
Keywords: biosynthetic gene cluster; conidia; hyphal growth; hypoxia; mycelial growth; specialized metabolism; srbA; velvet protein complex.
Figures




References
-
- Bennett JW, Bentley R. 1989. What’s in a name?—microbial secondary metabolism. Adv Appl Microbiol 34:1–28. doi:10.1016/S0065-2164(08)70316-2. - DOI
-
- Bayram Ö, Krappmann S, Ni M, Bok JW, Helmstaedt K, Yu J-H, Braus GH, Bayram O, Valerius O, Braus-Stromeyer S, Kwon N-J, Keller NP. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

