Improved Prediction of Blood-Brain Barrier Permeability Through Machine Learning with Combined Use of Molecular Property-Based Descriptors and Fingerprints
- PMID: 29564576
- PMCID: PMC7737623
- DOI: 10.1208/s12248-018-0215-8
Improved Prediction of Blood-Brain Barrier Permeability Through Machine Learning with Combined Use of Molecular Property-Based Descriptors and Fingerprints
Abstract
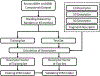
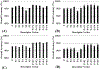
Blood-brain barrier (BBB) permeability of a compound determines whether the compound can effectively enter the brain. It is an essential property which must be accounted for in drug discovery with a target in the brain. Several computational methods have been used to predict the BBB permeability. In particular, support vector machine (SVM), which is a kernel-based machine learning method, has been used popularly in this field. For SVM training and prediction, the compounds are characterized by molecular descriptors. Some SVM models were based on the use of molecular property-based descriptors (including 1D, 2D, and 3D descriptors) or fragment-based descriptors (known as the fingerprints of a molecule). The selection of descriptors is critical for the performance of a SVM model. In this study, we aimed to develop a generally applicable new SVM model by combining all of the features of the molecular property-based descriptors and fingerprints to improve the accuracy for the BBB permeability prediction. The results indicate that our SVM model has improved accuracy compared to the currently available models of the BBB permeability prediction.
Keywords: blood–brain barrier permeability; fingerprint; modeling; molecular descriptor; physical property.
Conflict of interest statement
Competing Financial Interests statement: The authors declare that there is no conflict of interest for this work.
Figures
Similar articles
-
Uncovering blood-brain barrier permeability: a comparative study of machine learning models using molecular fingerprints, and SHAP explainability.SAR QSAR Environ Res. 2024 Dec;35(12):1155-1171. doi: 10.1080/1062936X.2024.2446352. Epub 2025 Jan 8. SAR QSAR Environ Res. 2024. PMID: 39773123
-
In Silico Prediction of Blood-Brain Barrier Permeability of Compounds by Machine Learning and Resampling Methods.ChemMedChem. 2018 Oct 22;13(20):2189-2201. doi: 10.1002/cmdc.201800533. Epub 2018 Sep 21. ChemMedChem. 2018. PMID: 30110511
-
A Genetic Algorithm Based Support Vector Machine Model for Blood-Brain Barrier Penetration Prediction.Biomed Res Int. 2015;2015:292683. doi: 10.1155/2015/292683. Epub 2015 Oct 4. Biomed Res Int. 2015. PMID: 26504797 Free PMC article.
-
Machine Learning in Drug Development for Neurological Diseases: A Review of Blood Brain Barrier Permeability Prediction Models.Mol Inform. 2025 Mar;44(3):e202400325. doi: 10.1002/minf.202400325. Mol Inform. 2025. PMID: 40146590 Free PMC article. Review.
-
Blood Brain Barrier Permeability Prediction Using Machine Learning Techniques: An Update.Curr Pharm Biotechnol. 2019;20(14):1163-1171. doi: 10.2174/1389201020666190821145346. Curr Pharm Biotechnol. 2019. PMID: 31433750 Review.
Cited by
-
Improving glioma drug delivery: A multifaceted approach for glioma drug development.Pharmacol Res. 2024 Oct;208:107390. doi: 10.1016/j.phrs.2024.107390. Epub 2024 Sep 2. Pharmacol Res. 2024. PMID: 39233056 Free PMC article. Review.
-
Multi-Targeting Approach in Glioblastoma Using Computer-Assisted Drug Discovery Tools to Overcome the Blood-Brain Barrier and Target EGFR/PI3Kp110β Signaling.Cancers (Basel). 2022 Jul 19;14(14):3506. doi: 10.3390/cancers14143506. Cancers (Basel). 2022. PMID: 35884571 Free PMC article.
-
Comparing the Pfizer Central Nervous System Multiparameter Optimization Calculator and a BBB Machine Learning Model.ACS Chem Neurosci. 2021 Jun 16;12(12):2247-2253. doi: 10.1021/acschemneuro.1c00265. Epub 2021 May 24. ACS Chem Neurosci. 2021. PMID: 34028255 Free PMC article.
-
Improved Classification of Blood-Brain-Barrier Drugs Using Deep Learning.Sci Rep. 2019 Jun 19;9(1):8802. doi: 10.1038/s41598-019-44773-4. Sci Rep. 2019. PMID: 31217424 Free PMC article.
-
A deep learning approach to predict blood-brain barrier permeability.PeerJ Comput Sci. 2021 Jun 10;7:e515. doi: 10.7717/peerj-cs.515. eCollection 2021. PeerJ Comput Sci. 2021. PMID: 34179448 Free PMC article.
References
-
- George A. The design and molecular modeling of CNS drugs. Current opinion in drug discovery & development. 1999;2(4):286–92. - PubMed
-
- Puzyn T, Leszczynski J, Cronin MT. Recent advances in QSAR studies: methods and applications: Springer Science & Business Media; 2010.
-
- Bicker J, Alves G, Fortuna A, Falcão A. Blood–brain barrier models and their relevance for a successful development of CNS drug delivery systems: a review. European Journal of Pharmaceutics and Biopharmaceutics. 2014;87(3):409–32. - PubMed
-
- Abraham MH. The factors that influence permeation across the blood–brain barrier. European journal of medicinal chemistry. 2004;39(3):235–40. - PubMed
-
- Rose K, Hall LH, Kier LB. Modeling blood-brain barrier partitioning using the electrotopological state. Journal of chemical information and computer sciences. 2002;42(3):651–66. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

