Negative Electron Transfer Dissociation Sequencing of 3-O-Sulfation-Containing Heparan Sulfate Oligosaccharides
- PMID: 29564812
- PMCID: PMC6004244
- DOI: 10.1007/s13361-018-1907-0
Negative Electron Transfer Dissociation Sequencing of 3-O-Sulfation-Containing Heparan Sulfate Oligosaccharides
Abstract
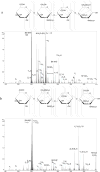
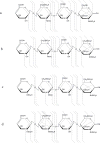
Among dissociation methods, negative electron transfer dissociation (NETD) has been proven the most useful for glycosaminoglycan (GAG) sequencing because it produces informative fragmentation, a low degree of sulfate losses, high sensitivity, and translatability to multiple instrument types. The challenge, however, is to distinguish positional sulfation. In particular, NETD has been reported to fail to differentiate 4-O- versus 6-O-sulfation in chondroitin sulfate decasaccharide. This raised the concern of whether NETD is able to differentiate the rare 3-O-sulfation from predominant 6-O-sulfation in heparan sulfate (HS) oligosaccharides. Here, we report that NETD generates highly informative spectra that differentiate sites of O-sulfation on glucosamine residues, enabling structural characterizations of synthetic HS isomers containing 3-O-sulfation. Further, lyase-resistant 3-O-sulfated tetrasaccharides from natural sources were successfully sequenced. Notably, for all of the oligosaccharides in this study, the successful sequencing is based on NETD tandem mass spectra of commonly observed deprotonated precursor ions without derivatization or metal cation adduction, simplifying the experimental workflow and data interpretation. These results demonstrate the potential of NETD as a sensitive analytical tool for detailed, high-throughput structural analysis of highly sulfated GAGs. Graphical Abstract.
Keywords: Fourier transform ion cyclotron resonance mass spectrometry; Glycomics; Glycosaminoglycan; Heparan sulfate; Negative electron transfer dissociation; Sulfation.
Figures




Similar articles
-
Sequencing Heparan Sulfate Using HILIC LC-NETD-MS/MS.Anal Chem. 2019 Sep 17;91(18):11738-11746. doi: 10.1021/acs.analchem.9b02313. Epub 2019 Aug 29. Anal Chem. 2019. PMID: 31423779 Free PMC article.
-
De novo sequencing of heparan sulfate oligosaccharides by electron-activated dissociation.Anal Chem. 2013 Dec 17;85(24):11979-86. doi: 10.1021/ac402931j. Epub 2013 Nov 22. Anal Chem. 2013. PMID: 24224699 Free PMC article.
-
Distinguishing glucuronic from iduronic acid in glycosaminoglycan tetrasaccharides by using electron detachment dissociation.Anal Chem. 2007 Mar 1;79(5):2015-22. doi: 10.1021/ac061636x. Epub 2007 Jan 25. Anal Chem. 2007. PMID: 17253657 Free PMC article.
-
Recent advances in the synthesis of extensive libraries of heparan sulfate oligosaccharides for structure-activity relationship studies.Curr Opin Chem Biol. 2024 Jun;80:102455. doi: 10.1016/j.cbpa.2024.102455. Epub 2024 Apr 17. Curr Opin Chem Biol. 2024. PMID: 38636446 Free PMC article. Review.
-
Chemoenzymatic synthesis of glycosaminoglycans: re-creating, re-modeling and re-designing nature's longest or most complex carbohydrate chains.Glycobiology. 2013 Jul;23(7):764-77. doi: 10.1093/glycob/cwt016. Epub 2013 Mar 11. Glycobiology. 2013. PMID: 23481097 Free PMC article. Review.
Cited by
-
Mass Spectrometry-Based Techniques to Elucidate the Sugar Code.Chem Rev. 2022 Apr 27;122(8):7840-7908. doi: 10.1021/acs.chemrev.1c00380. Epub 2021 Sep 7. Chem Rev. 2022. PMID: 34491038 Free PMC article. Review.
-
Developments in Mass Spectrometry for Glycosaminoglycan Analysis: A Review.Mol Cell Proteomics. 2021;20:100025. doi: 10.1074/mcp.R120.002267. Epub 2021 Jan 6. Mol Cell Proteomics. 2021. PMID: 32938749 Free PMC article. Review.
-
Influence of saccharide modifications on heparin lyase III substrate specificities.Glycobiology. 2022 Mar 30;32(3):208-217. doi: 10.1093/glycob/cwab023. Glycobiology. 2022. PMID: 33822051 Free PMC article.
-
The 3-O-sulfation of heparan sulfate modulates protein binding and lyase degradation.Proc Natl Acad Sci U S A. 2021 Jan 19;118(3):e2012935118. doi: 10.1073/pnas.2012935118. Proc Natl Acad Sci U S A. 2021. PMID: 33441484 Free PMC article.
-
Alteration of Neuropilin-1 and Heparan Sulfate Interaction Impairs Murine B16 Tumor Growth.ACS Chem Biol. 2024 Aug 16;19(8):1820-1835. doi: 10.1021/acschembio.4c00389. Epub 2024 Aug 4. ACS Chem Biol. 2024. PMID: 39099090 Free PMC article.
References
-
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–71. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

