Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits
- PMID: 29565274
- PMCID: PMC6023089
- DOI: 10.3390/antibiotics7020028
Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits
Abstract
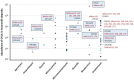
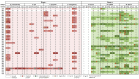
Cave moonmilk deposits host an abundant and diverse actinobacterial population that has a great potential for producing novel natural bioactive compounds. In our previous attempt to isolate culturable moonmilk-dwelling Actinobacteria, only Streptomyces species were recovered, whereas a metagenetic study of the same deposits revealed a complex actinobacterial community including 46 actinobacterial genera in addition to streptomycetes. In this work, we applied the rehydration-centrifugation method to lessen the occurrence of filamentous species and tested a series of strategies to achieve the isolation of hard-to-culture and rare Actinobacteria from the moonmilk deposits of the cave "Grotte des Collemboles". From the "tips and tricks" that were tested, separate autoclaving of the components of the International Streptomyces Project (ISP) medium number 5 (ISP5) medium, prolonged incubation time, and dilution of the moonmilk suspension were found to most effectively improve colony forming units. Taxonomic analyses of the 40 isolates revealed new representatives of the Agromyces, Amycolatopsis, Kocuria, Micrococcus, Micromonospora, Nocardia, and Rhodococcus species, as well as additional new streptomycetes. The applied methodologies allowed the isolation of strains associated with both the least and most abundant moonmilk-dwelling actinobacterial operational taxonomic units. Finally, bioactivity screenings revealed that some isolates displayed high antibacterial activities, and genome mining uncovered a strong potential for the production of natural compounds.
Keywords: Amycolatopsis; Cave microbiology; Streptomyces; antibiotics; rare Actinobacteria; secondary metabolite; siderophore; unculturability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- World Health Organization (WHO) Antimicrobial Resistance: Global Report on Surveillance 2014. WHO; Geneva, Switzerland: 2014. AMR Report.
-
- Hutchinson G.E. The Paradox of the Plankton. Am. Nat. 1961;95:137–145. doi: 10.1086/282171. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

