Next-generation sequencing analysis of a cluster of hepatitis C virus infections in a haematology and oncology center
- PMID: 29566084
- PMCID: PMC5864040
- DOI: 10.1371/journal.pone.0194816
Next-generation sequencing analysis of a cluster of hepatitis C virus infections in a haematology and oncology center
Abstract
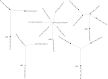
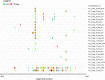
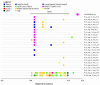
Molecular characterization of early hepatitis C virus (HCV) infection remains rare. Ten out of 78 patients of a hematology/oncology center were found to be HCV RNA positive two to four months after hospitalization. Only two of the ten patients were anti-HCV positive. HCV hypervariable region 1 (HVR1) was amplified in seven patients (including one anti-HCV positive) and analyzed by next generation sequencing (NGS). Genetic variants were reconstructed by Shorah and an empirically established 0.5% variant frequency cut-off was implemented. These sequences were compared by phylogenetic and diversity analyses. Ten unrelated blood donors with newly acquired HCV infection detected at the time of donation (HCV RNA positive and anti-HCV negative) served as controls. One to seven HVR1 variants were found in each patient. Sequences intermixed phylogenetically with no evidence of clustering in individual patients. These sequences were more similar to each other (similarity 95.4% to 100.0%) than to those of controls (similarity 64.8% to 82.6%). An identical predominant variant was present in four patients, whereas other closely related variants dominated in the remaining three patients. In five patients the HCV population was limited to a single variant or one predominant variant and minor variants of less than 10% frequency. In conclusion, NGS analysis of a cluster of HCV infections acquired in the hospital setting revealed the presence of low diversity, very closely related variants in all patients, suggesting an early-stage infection with the same virus. NGS combined with phylogenetic analysis and classical epidemiological analysis could help in tracking of HCV outbreaks.
Conflict of interest statement
Figures


References
-
- Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288(5464):339–44. . - PubMed
-
- Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiology and molecular biology reviews: MMBR. 2012;76(2):159–216. doi: 10.1128/MMBR.05023-11 ; PubMed Central PMCID: PMC3372249. - DOI - PMC - PubMed
-
- Wang GP, Sherrill-Mix SA, Chang KM, Quince C, Bushman FD. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J Virol. 2010;84(12):6218–28. doi: 10.1128/JVI.02271-09 ; PubMed Central PMCID: PMCPMC2876626. - DOI - PMC - PubMed
-
- Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, et al. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS pathogens. 2011;7(9):e1002243 doi: 10.1371/journal.ppat.1002243 ; PubMed Central PMCID: PMCPMC3164670. - DOI - PMC - PubMed
-
- Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, et al. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS pathogens. 2012;8(8):e1002880 doi: 10.1371/journal.ppat.1002880 ; PubMed Central PMCID: PMCPMC3426529. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

