Biomedical informatics and machine learning for clinical genomics
- PMID: 29566172
- PMCID: PMC5946905
- DOI: 10.1093/hmg/ddy088
Biomedical informatics and machine learning for clinical genomics
Abstract
While tens of thousands of pathogenic variants are used to inform the many clinical applications of genomics, there remains limited information on quantitative disease risk for the majority of variants used in clinical practice. At the same time, rising demand for genetic counselling has prompted a growing need for computational approaches that can help interpret genetic variation. Such tasks include predicting variant pathogenicity and identifying variants that are too common to be penetrant. To address these challenges, researchers are increasingly turning to integrative informatics approaches. These approaches often leverage vast sources of data, including electronic health records and population-level allele frequency databases (e.g. gnomAD), as well as machine learning techniques such as support vector machines and deep learning. In this review, we highlight recent informatics and machine learning approaches that are improving our understanding of pathogenic variation and discuss obstacles that may limit their emerging role in clinical genomics.
Figures
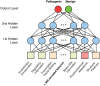
References
-
- Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet., 12, 745–755. - PubMed
-
- Manrai A.K., Ioannidis J.P.A., Kohane I.S. (2016) Clinical genomics: from pathogenicity claims to quantitative risk estimates. JAMA, 315, 1233–1234. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

