PHD-finger domain protein 5A functions as a novel oncoprotein in lung adenocarcinoma
- PMID: 29566713
- PMCID: PMC5863814
- DOI: 10.1186/s13046-018-0736-0
PHD-finger domain protein 5A functions as a novel oncoprotein in lung adenocarcinoma
Abstract
Background: PHD-finger domain protein 5A (PHF5A) is a highly conserved small transcriptional regulator also involved in pre-mRNA splicing; however, its biological functions and molecular mechanisms in non-small cell lung cancer (NSCLC) have not yet been investigated. The purpose of this study was to determine the functional relevance and therapeutic potential of PHF5A in lung adenocarcinoma (LAC).
Methods: The expression of PHF5A in LAC tissues and adjacent non-tumor (ANT) tissues was investigated using immunohistochemistry of a tissue microarray, qRT-PCR, western blot and bioinformatics. The function of PHF5A was determined using several in vitro assays and also in vivo assay by lentiviral vector-mediated PHF5A depletion in LAC cell lines.
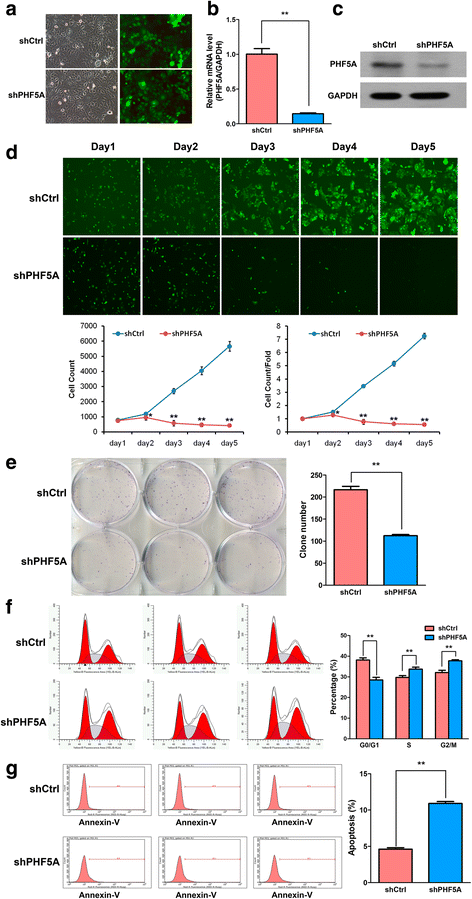
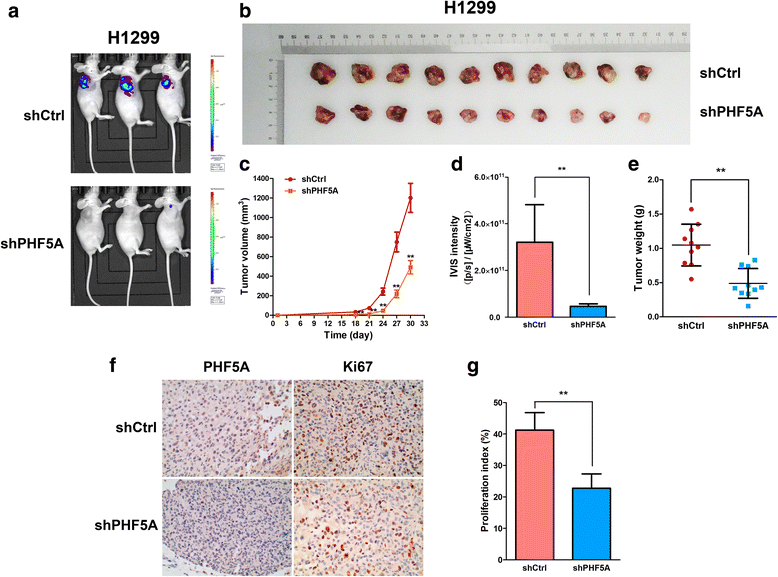
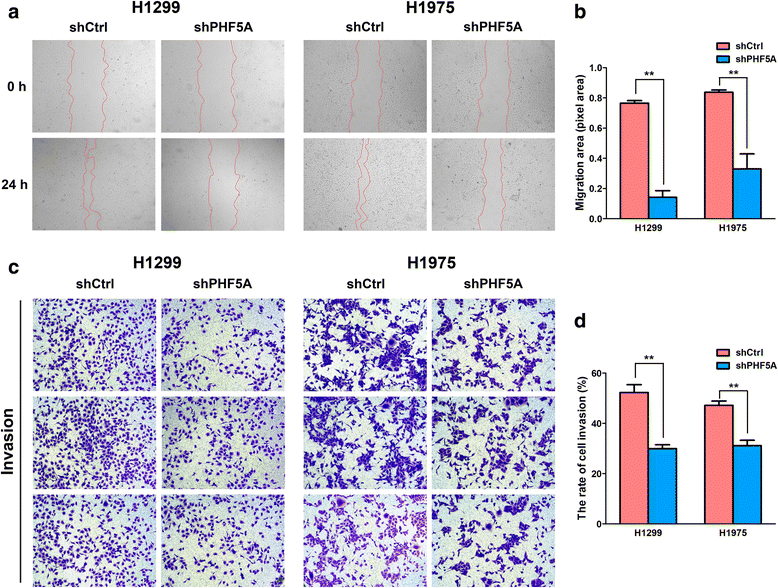
Results: PHF5A was highly upregulated in LAC tissues compared with the ANT counterparts, and closely associated with tumor progression and poor patient prognosis. These results were further confirmed by findings of the TCGA database. Moreover, functional studies demonstrated that PHF5A knockdown not only resulted in reduced cell proliferation, increased cell apoptosis, and cell cycle arrest, but also suppressed migration and invasion in LAC cells. PHF5A silencing was also found to inhibit LAC tumor growth in nude mice. Microarray and bioinformatics analyses revealed that PHF5A depletion led to dysregulation of multiple tumor signaling pathways; selected factors in key signaling pathways were verified in vitro.
Conclusions: The data suggest for the first time that PHF5A is an oncoprotein that contributes to LAC progression by regulating multiple signaling pathways, and may constitute a prognostic factor and potential new therapeutic target in NSCLC.
Keywords: Lung adenocarcinoma; PHF5A; Prognostic biomarker; Proliferation; Tumor invasion.
Conflict of interest statement
Ethics approval
All animal experiments were performed according to national guidelines and approved by the Animal Care and Use Ethics Committee of Bengbu Medical College (Bengbu, China). Approval for clinical sample collection was obtained from the medical ethics committee of our institute, and written informed consent was provided by all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
PHF5A as a new OncoTarget and therapeutic prospects.Heliyon. 2023 Jul 6;9(7):e18010. doi: 10.1016/j.heliyon.2023.e18010. eCollection 2023 Jul. Heliyon. 2023. PMID: 37483794 Free PMC article. Review.
-
PHD finger protein 5A promoted lung adenocarcinoma progression via alternative splicing.Cancer Med. 2019 May;8(5):2429-2441. doi: 10.1002/cam4.2115. Epub 2019 Apr 1. Cancer Med. 2019. PMID: 30932358 Free PMC article.
-
PHF5A Epigenetically Inhibits Apoptosis to Promote Breast Cancer Progression.Cancer Res. 2018 Jun 15;78(12):3190-3206. doi: 10.1158/0008-5472.CAN-17-3514. Epub 2018 Apr 26. Cancer Res. 2018. PMID: 29700004
-
SOX30 is a key regulator of desmosomal gene suppressing tumor growth and metastasis in lung adenocarcinoma.J Exp Clin Cancer Res. 2018 May 31;37(1):111. doi: 10.1186/s13046-018-0778-3. J Exp Clin Cancer Res. 2018. PMID: 29855376 Free PMC article.
-
Research progress and therapeutic prospect of PHF5A acting as a new target for malignant tumors.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022 Nov 25;51(5):647-655. doi: 10.3724/zdxbyxb-2022-0459. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022. PMID: 36581580 Free PMC article. Review. English.
Cited by
-
Comprehensive analysis of PHF5A as a potential prognostic biomarker and therapeutic target across cancers and in hepatocellular carcinoma.BMC Cancer. 2024 Jul 19;24(1):868. doi: 10.1186/s12885-024-12620-z. BMC Cancer. 2024. PMID: 39030507 Free PMC article.
-
MicroRNA-302a/d inhibits the self-renewal capability and cell cycle entry of liver cancer stem cells by targeting the E2F7/AKT axis.J Exp Clin Cancer Res. 2018 Oct 16;37(1):252. doi: 10.1186/s13046-018-0927-8. J Exp Clin Cancer Res. 2018. PMID: 30326936 Free PMC article.
-
The SF3b complex: splicing and beyond.Cell Mol Life Sci. 2020 Sep;77(18):3583-3595. doi: 10.1007/s00018-020-03493-z. Epub 2020 Mar 5. Cell Mol Life Sci. 2020. PMID: 32140746 Free PMC article. Review.
-
PHF5A as a new OncoTarget and therapeutic prospects.Heliyon. 2023 Jul 6;9(7):e18010. doi: 10.1016/j.heliyon.2023.e18010. eCollection 2023 Jul. Heliyon. 2023. PMID: 37483794 Free PMC article. Review.
-
Knockdown of PHF5A Inhibits Migration and Invasion of HCC Cells via Downregulating NF-κB Signaling.Biomed Res Int. 2019 Jan 15;2019:1621854. doi: 10.1155/2019/1621854. eCollection 2019. Biomed Res Int. 2019. PMID: 30766880 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

