Discovery of a novel allosteric inhibitor scaffold for polyadenosine-diphosphate-ribose polymerase 14 (PARP14) macrodomain 2
- PMID: 29567296
- PMCID: PMC6008491
- DOI: 10.1016/j.bmc.2018.03.020
Discovery of a novel allosteric inhibitor scaffold for polyadenosine-diphosphate-ribose polymerase 14 (PARP14) macrodomain 2
Abstract
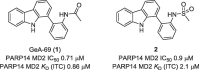
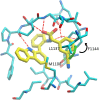
The polyadenosine-diphosphate-ribose polymerase 14 (PARP14) has been implicated in DNA damage response pathways for homologous recombination. PARP14 contains three (ADP ribose binding) macrodomains (MD) whose exact contribution to overall PARP14 function in pathology remains unclear. A medium throughput screen led to the identification of N-(2(-9H-carbazol-1-yl)phenyl)acetamide (GeA-69, 1) as a novel allosteric PARP14 MD2 (second MD of PARP14) inhibitor. We herein report medicinal chemistry around this novel chemotype to afford a sub-micromolar PARP14 MD2 inhibitor. This chemical series provides a novel starting point for further development of PARP14 chemical probes.
Keywords: Inhibitor Design; Macrodomain; PARP; Poly-ADP ribsose.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures












References
-
- Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. - PubMed
- Rack J.G.M., Perina D., Ahel I. Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu Rev Biochem. 2016;85:431–454. - PubMed
- Wahlberg E., Karlberg T., Kouznetsova E. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotech. 2012;30:283–288. - PubMed
-
- Kleine H., Poreba E., Lesniewicz K. Substrate-Assisted Catalysis by PARP10 Limits Its Activity to Mono-ADP-Ribosylation. Mol Cell. 2008;32:57–69. - PubMed
- Ame J.-C., Spenlehauer C., de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. - PubMed
- Vyas S., Matic I., Uchima L. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. - PMC - PubMed
-
- Hottiger M.O., Hassa P.O., Lüscher B., Schüler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

