Pan-SRC kinase inhibition blocks B-cell receptor oncogenic signaling in non-Hodgkin lymphoma
- PMID: 29567799
- PMCID: PMC5969377
- DOI: 10.1182/blood-2017-10-809210
Pan-SRC kinase inhibition blocks B-cell receptor oncogenic signaling in non-Hodgkin lymphoma
Abstract
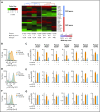
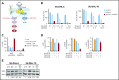
In diffuse large B-cell lymphoma (DLBCL), activation of the B-cell receptor (BCR) promotes multiple oncogenic signals, which are essential for tumor proliferation. Inhibition of the Bruton's tyrosine kinase (BTK), a BCR downstream target, is therapeutically effective only in a subgroup of patients with DLBCL. Here, we used lymphoma cells isolated from patients with DLBCL to measure the effects of targeted therapies on BCR signaling and to anticipate response. In lymphomas resistant to BTK inhibition, we show that blocking BTK activity enhanced tumor dependencies from alternative oncogenic signals downstream of the BCR, converging on MYC upregulation. To completely ablate the activity of the BCR, we genetically and pharmacologically repressed the activity of the SRC kinases LYN, FYN, and BLK, which are responsible for the propagation of the BCR signal. Inhibition of these kinases strongly reduced tumor growth in xenografts and cell lines derived from patients with DLBCL independent of their molecular subtype, advancing the possibility to be relevant therapeutic targets in broad and diverse groups of DLBCL patients.
© 2018 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures





Comment in
-
The future of kinase inhibitors for DLBCL?Blood. 2018 May 24;131(21):2278-2280. doi: 10.1182/blood-2018-04-841908. Blood. 2018. PMID: 29794076 Free PMC article.
References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, et al. ; Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937-1947. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

