Engineering enhanced cellobiohydrolase activity
- PMID: 29567941
- PMCID: PMC5864845
- DOI: 10.1038/s41467-018-03501-8
Engineering enhanced cellobiohydrolase activity
Abstract
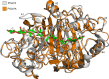
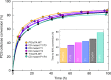
Glycoside Hydrolase Family 7 cellobiohydrolases (GH7 CBHs) catalyze cellulose depolymerization in cellulolytic eukaryotes, making them key discovery and engineering targets. However, there remains a lack of robust structure-activity relationships for these industrially important cellulases. Here, we compare CBHs from Trichoderma reesei (TrCel7A) and Penicillium funiculosum (PfCel7A), which exhibit a multi-modular architecture consisting of catalytic domain (CD), carbohydrate-binding module, and linker. We show that PfCel7A exhibits 60% greater performance on biomass than TrCel7A. To understand the contribution of each domain to this improvement, we measure enzymatic activity for a library of CBH chimeras with swapped subdomains, demonstrating that the enhancement is mainly caused by PfCel7A CD. We solve the crystal structure of PfCel7A CD and use this information to create a second library of TrCel7A CD mutants, identifying a TrCel7A double mutant with near-equivalent activity to wild-type PfCel7A. Overall, these results reveal CBH regions that enable targeted activity improvements.
Conflict of interest statement
W.S.A., J.O.B., S.R.D., M.E.H., G.T.B., L.E.T., J.G.L., K.P., and Q.X. hold a patent related to
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

