Dual binding in cohesin-dockerin complexes: the energy landscape and the role of short, terminal segments of the dockerin module
- PMID: 29568013
- PMCID: PMC5864761
- DOI: 10.1038/s41598-018-23380-9
Dual binding in cohesin-dockerin complexes: the energy landscape and the role of short, terminal segments of the dockerin module
Abstract
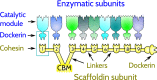
The assembly of the polysaccharide degradating cellulosome machinery is mediated by tight binding between cohesin and dockerin domains. We have used an empirical model known as FoldX as well as molecular mechanics methods to determine the free energy of binding between a cohesin and a dockerin from Clostridium thermocellum in two possible modes that differ by an approximately 180° rotation. Our studies suggest that the full-length wild-type complex exhibits dual binding at room temperature, i.e., the two modes of binding have comparable probabilities at equilibrium. The ability to bind in the two modes persists at elevated temperatures. However, single-point mutations or truncations of terminal segments in the dockerin result in shifting the equilibrium towards one of the binding modes. Our molecular dynamics simulations of mechanical stretching of the full-length wild-type cohesin-dockerin complex indicate that each mode of binding leads to two kinds of stretching pathways, which may be mistakenly taken as evidence of dual binding.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Novel Clostridium thermocellum type I cohesin-dockerin complexes reveal a single binding mode.J Biol Chem. 2012 Dec 28;287(53):44394-405. doi: 10.1074/jbc.M112.407700. Epub 2012 Nov 1. J Biol Chem. 2012. PMID: 23118225 Free PMC article.
-
Probing the mechanism of cellulosome attachment to the Clostridium thermocellum cell surface: computer simulation of the Type II cohesin-dockerin complex and its variants.Protein Eng Des Sel. 2010 Oct;23(10):759-68. doi: 10.1093/protein/gzq049. Epub 2010 Aug 3. Protein Eng Des Sel. 2010. PMID: 20682763
-
Dynamic interactions of type I cohesin modules fine-tune the structure of the cellulosome of Clostridium thermocellum.Proc Natl Acad Sci U S A. 2018 Nov 27;115(48):E11274-E11283. doi: 10.1073/pnas.1809283115. Epub 2018 Nov 14. Proc Natl Acad Sci U S A. 2018. PMID: 30429330 Free PMC article.
-
Insights into cellulosome assembly and dynamics: from dissection to reconstruction of the supramolecular enzyme complex.Curr Opin Struct Biol. 2013 Oct;23(5):686-94. doi: 10.1016/j.sbi.2013.09.002. Epub 2013 Sep 27. Curr Opin Struct Biol. 2013. PMID: 24080387 Review.
-
Single versus dual-binding conformations in cellulosomal cohesin-dockerin complexes.Curr Opin Struct Biol. 2016 Oct;40:89-96. doi: 10.1016/j.sbi.2016.08.002. Epub 2016 Aug 28. Curr Opin Struct Biol. 2016. PMID: 27579515 Review.
Cited by
-
Nanoscale resolution of microbial fiber degradation in action.Elife. 2022 May 31;11:e76523. doi: 10.7554/eLife.76523. Elife. 2022. PMID: 35638899 Free PMC article.
-
In silico characterization of the GH5-cellulase family from uncultured microorganisms: physicochemical and structural studies.J Genet Eng Biotechnol. 2021 Sep 30;19(1):143. doi: 10.1186/s43141-021-00236-w. J Genet Eng Biotechnol. 2021. PMID: 34591195 Free PMC article.
-
Enzymatic degradation of cellulose in soil: A review.Heliyon. 2024 Jan 3;10(1):e24022. doi: 10.1016/j.heliyon.2024.e24022. eCollection 2024 Jan 15. Heliyon. 2024. PMID: 38234915 Free PMC article.
-
Current challenges in designer cellulosome engineering.Appl Microbiol Biotechnol. 2023 May;107(9):2755-2770. doi: 10.1007/s00253-023-12474-8. Epub 2023 Mar 21. Appl Microbiol Biotechnol. 2023. PMID: 36941434 Review.
References
-
- Beguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 1996;31:201236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

