An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations
- PMID: 29568061
- PMCID: PMC5864832
- DOI: 10.1038/s41467-018-03523-2
An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations
Abstract
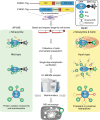
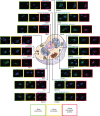
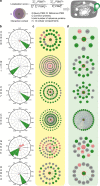
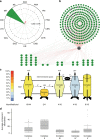
Protein-protein interactions govern almost all cellular functions. These complex networks of stable and transient associations can be mapped by affinity purification mass spectrometry (AP-MS) and complementary proximity-based labeling methods such as BioID. To exploit the advantages of both strategies, we here design and optimize an integrated approach combining AP-MS and BioID in a single construct, which we term MAC-tag. We systematically apply the MAC-tag approach to 18 subcellular and 3 sub-organelle localization markers, generating a molecular context database, which can be used to define a protein's molecular location. In addition, we show that combining the AP-MS and BioID results makes it possible to obtain interaction distances within a protein complex. Taken together, our integrated strategy enables the comprehensive mapping of the physical and functional interactions of proteins, defining their molecular context and improving our understanding of the cellular interactome.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Combined proximity labeling and affinity purification-mass spectrometry workflow for mapping and visualizing protein interaction networks.Nat Protoc. 2020 Oct;15(10):3182-3211. doi: 10.1038/s41596-020-0365-x. Epub 2020 Aug 10. Nat Protoc. 2020. PMID: 32778839
-
Parallel Exploration of Interaction Space by BioID and Affinity Purification Coupled to Mass Spectrometry.Methods Mol Biol. 2017;1550:115-136. doi: 10.1007/978-1-4939-6747-6_10. Methods Mol Biol. 2017. PMID: 28188527
-
Analyzing Protein Interactions by MAC-Tag Approaches.Methods Mol Biol. 2023;2690:281-297. doi: 10.1007/978-1-0716-3327-4_24. Methods Mol Biol. 2023. PMID: 37450155
-
Proteomic navigation using proximity-labeling.Methods. 2019 Jul 15;164-165:67-72. doi: 10.1016/j.ymeth.2019.03.028. Epub 2019 Apr 4. Methods. 2019. PMID: 30953756 Review.
-
Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles.Curr Opin Chem Biol. 2019 Feb;48:44-54. doi: 10.1016/j.cbpa.2018.10.017. Epub 2018 Nov 17. Curr Opin Chem Biol. 2019. PMID: 30458335 Review.
Cited by
-
Circ-TRIO promotes TNBC progression by regulating the miR-432-5p/CCDC58 axis.Cell Death Dis. 2022 Sep 8;13(9):776. doi: 10.1038/s41419-022-05216-7. Cell Death Dis. 2022. PMID: 36075896 Free PMC article.
-
Novel TMEM173 Mutation and the Role of Disease Modifying Alleles.Front Immunol. 2019 Dec 5;10:2770. doi: 10.3389/fimmu.2019.02770. eCollection 2019. Front Immunol. 2019. PMID: 31866997 Free PMC article.
-
Proximity-Dependent Biotinylation Approaches to Explore the Dynamic Compartmentalized Proteome.Front Mol Biosci. 2022 Mar 4;9:852911. doi: 10.3389/fmolb.2022.852911. eCollection 2022. Front Mol Biosci. 2022. PMID: 35309513 Free PMC article. Review.
-
A partnership between the lipid scramblase XK and the lipid transfer protein VPS13A at the plasma membrane.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2205425119. doi: 10.1073/pnas.2205425119. Epub 2022 Aug 22. Proc Natl Acad Sci U S A. 2022. PMID: 35994651 Free PMC article.
-
Nuclear actin interactome analysis links actin to KAT14 histone acetyl transferase and mRNA splicing.J Cell Sci. 2019 Apr 17;132(8):jcs226852. doi: 10.1242/jcs.226852. J Cell Sci. 2019. PMID: 30890647 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

