Population genomic analysis of the rice blast fungus reveals specific events associated with expansion of three main clades
- PMID: 29568114
- PMCID: PMC6051997
- DOI: 10.1038/s41396-018-0100-6
Population genomic analysis of the rice blast fungus reveals specific events associated with expansion of three main clades
Abstract
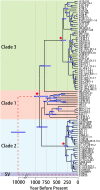
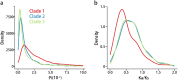
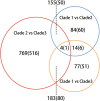
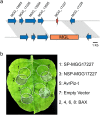
We examined the genomes of 100 isolates of Magnaporthe oryzae (Pyricularia oryzae), the causal agent of rice blast disease. We grouped current field populations of M. oryzae into three major globally distributed groups. A genetically diverse group, clade 1, which may represent a group of closely related lineages, contains isolates of both mating types. Two well-separated clades, clades 2 and 3, appear to have arisen as clonal lineages distinct from the genetically diverse clade. Examination of genes involved in mating pathways identified clade-specific diversification of several genes with orthologs involved in mating behavior in other fungi. All isolates within each clonal lineage are of the same mating type. Clade 2 is distinguished by a unique deletion allele of a gene encoding a small cysteine-rich protein that we determined to be a virulence factor. Clade 3 isolates have a small deletion within the MFA2 pheromone precursor gene, and this allele is shared with an unusual group of isolates we placed within clade 1 that contain AVR1-CO39 alleles. These markers could be used for rapid screening of isolates and suggest specific events in evolution that shaped these populations. Our findings are consistent with the view that M. oryzae populations in Asia generate diversity through recombination and may have served as the source of the clades 2 and 3 isolates that comprise a large fraction of the global population.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Coexistence of Multiple Endemic and Pandemic Lineages of the Rice Blast Pathogen.mBio. 2018 Apr 3;9(2):e01806-17. doi: 10.1128/mBio.01806-17. mBio. 2018. PMID: 29615506 Free PMC article.
-
Deciphering Genome Content and Evolutionary Relationships of Isolates from the Fungus Magnaporthe oryzae Attacking Different Host Plants.Genome Biol Evol. 2015 Oct 9;7(10):2896-912. doi: 10.1093/gbe/evv187. Genome Biol Evol. 2015. PMID: 26454013 Free PMC article.
-
Transposable elements impact the population divergence of rice blast fungus Magnaporthe oryzae.mBio. 2024 May 8;15(5):e0008624. doi: 10.1128/mbio.00086-24. Epub 2024 Mar 27. mBio. 2024. PMID: 38534157 Free PMC article.
-
Functional genomics in the rice blast fungus to unravel the fungal pathogenicity.J Zhejiang Univ Sci B. 2008 Oct;9(10):747-52. doi: 10.1631/jzus.B0860014. J Zhejiang Univ Sci B. 2008. PMID: 18837101 Free PMC article. Review.
-
The Role of Cell Wall Degrading Enzymes in Pathogenesis of Magnaporthe oryzae.Curr Protein Pept Sci. 2017;18(10):1019-1034. doi: 10.2174/1389203717666160813164955. Curr Protein Pept Sci. 2017. PMID: 27526928 Review.
Cited by
-
Contribution of recent technological advances to future resistance breeding.Theor Appl Genet. 2019 Mar;132(3):713-732. doi: 10.1007/s00122-019-03297-1. Epub 2019 Feb 12. Theor Appl Genet. 2019. PMID: 30756126 Review.
-
Emergence and spread of the barley net blotch pathogen coincided with crop domestication and cultivation history.PLoS Genet. 2024 Jan 29;20(1):e1010884. doi: 10.1371/journal.pgen.1010884. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38285729 Free PMC article.
-
Distinct genomic contexts predict gene presence-absence variation in different pathotypes of Magnaporthe oryzae.Genetics. 2024 Apr 3;226(4):iyae012. doi: 10.1093/genetics/iyae012. Genetics. 2024. PMID: 38290434 Free PMC article.
-
Differential loss of effector genes in three recently expanded pandemic clonal lineages of the rice blast fungus.BMC Biol. 2020 Jul 16;18(1):88. doi: 10.1186/s12915-020-00818-z. BMC Biol. 2020. PMID: 32677941 Free PMC article.
-
A fungal core effector exploits the OsPUX8B.2-OsCDC48-6 module to suppress plant immunity.Nat Commun. 2024 Mar 22;15(1):2559. doi: 10.1038/s41467-024-46903-7. Nat Commun. 2024. PMID: 38519521 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

