Effects of landscape features on population genetic variation of a tropical stream fish, Stone lapping minnow, Garra cambodgiensis, in the upper Nan River drainage basin, northern Thailand
- PMID: 29568710
- PMCID: PMC5845392
- DOI: 10.7717/peerj.4487
Effects of landscape features on population genetic variation of a tropical stream fish, Stone lapping minnow, Garra cambodgiensis, in the upper Nan River drainage basin, northern Thailand
Abstract
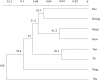
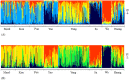
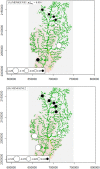
Spatial genetic variation of river-dwelling freshwater fishes is typically affected by the historical and contemporary river landscape as well as life-history traits. Tropical river and stream landscapes have endured extended geological change, shaping the existing pattern of genetic diversity, but were not directly affected by glaciation. Thus, spatial genetic variation of tropical fish populations should look very different from the pattern observed in temperate fish populations. These data are becoming important for designing appropriate management and conservation plans, as these aquatic systems are undergoing intense development and exploitation. This study evaluated the effects of landscape features on population genetic diversity of Garra cambodgiensis, a stream cyprinid, in eight tributary streams in the upper Nan River drainage basin (n = 30-100 individuals/location), Nan Province, Thailand. These populations are under intense fishing pressure from local communities. Based on 11 microsatellite loci, we detected moderate genetic diversity within eight population samples (average number of alleles per locus = 10.99 ± 3.00; allelic richness = 10.12 ± 2.44). Allelic richness within samples and stream order of the sampling location were negatively correlated (P < 0.05). We did not detect recent bottleneck events in these populations, but we did detect genetic divergence among populations (Global FST = 0.022, P < 0.01). The Bayesian clustering algorithms (TESS and STRUCTURE) suggested that four to five genetic clusters roughly coincide with sub-basins: (1) headwater streams/main stem of the Nan River, (2) a middle tributary, (3) a southeastern tributary and (4) a southwestern tributary. We observed positive correlation between geographic distance and linearized FST (P < 0.05), and the genetic differentiation pattern can be moderately explained by the contemporary stream network (STREAMTREE analysis, R2 = 0.75). The MEMGENE analysis suggested genetic division between northern (genetic clusters 1 and 2) and southern (clusters 3 and 4) sub-basins. We observed a high degree of genetic admixture in each location, highlighting the importance of natural flooding patterns and possible genetic impacts of supplementary stocking. Insights obtained from this research advance our knowledge of the complexity of a tropical stream system, and guide current conservation and restoration efforts for this species in Thailand.
Keywords: Garra cambodgiensis; Landscape genetics; Microsatellite variation; Spatial genetic variation; Tropical stream fish; Upper Nan River.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

Similar articles
-
The largest fish in the world's biggest river: Genetic connectivity and conservation of Arapaima gigas in the Amazon and Araguaia-Tocantins drainages.PLoS One. 2019 Aug 16;14(8):e0220882. doi: 10.1371/journal.pone.0220882. eCollection 2019. PLoS One. 2019. PMID: 31419237 Free PMC article.
-
Broad-scale sampling of primary freshwater fish populations reveals the role of intrinsic traits, inter-basin connectivity, drainage area and latitude on shaping contemporary patterns of genetic diversity.PeerJ. 2016 Feb 29;4:e1694. doi: 10.7717/peerj.1694. eCollection 2016. PeerJ. 2016. PMID: 26966653 Free PMC article.
-
Garra waensis, a new cyprinid fish (Actinopterygii: Cypriniformes) from the Nan River Basin of the Chao Phraya River system, northern Thailand.Zootaxa. 2014 Apr 23;3790:543-54. doi: 10.11646/zootaxa.3790.4.3. Zootaxa. 2014. PMID: 24869886
-
Fish faunal provinces of the conterminous United States of America reflect historical geography and familial composition.Biol Rev Camb Philos Soc. 2016 Aug;91(3):813-32. doi: 10.1111/brv.12196. Epub 2015 May 29. Biol Rev Camb Philos Soc. 2016. PMID: 26031190 Review.
-
A comprehensive review of the biodiversity of freshwater fish species in Valleys worldwide and in the Kingdom of Saudi Arabia.J Adv Vet Anim Res. 2024 Jun 8;11(2):356-366. doi: 10.5455/javar.2024.k784. eCollection 2024 Jun. J Adv Vet Anim Res. 2024. PMID: 39101086 Free PMC article. Review.
Cited by
-
Population genetics of the African snakehead fish Parachanna obscura along West Africa's water networks: Implications for sustainable management and conservation.Ecol Evol. 2023 Jan 16;13(1):e9724. doi: 10.1002/ece3.9724. eCollection 2023 Jan. Ecol Evol. 2023. PMID: 36694547 Free PMC article.
-
Assessing Genetic Variation and Population Connectivity in Wallago attu: A Call for Enhanced Conservation Measures.Biochem Genet. 2025 Jun 12. doi: 10.1007/s10528-025-11156-6. Online ahead of print. Biochem Genet. 2025. PMID: 40504468
References
-
- Allendorf FW, Luikart G. Conservation and the genetics of populations. Blackwell Publishing; Malden: 2007.
-
- Apodaca JJ, Rissler LJ, Godwin JC. Population structure and gene flow in a heavily disturbed habitat: implications for the management of the imperiled Red Hills salamander (Phaeognathus hubrichti) Conservation Genetic Resources. 2012;13(4):913–923. doi: 10.1007/s10592-012-0340-3. - DOI
-
- Barson NJ, Cable J, Van Oosterhout C. Population genetic analysis of microsatellite variation of guppies (Poecilia reticulata) in Trinidad and Tobago: evidence for a dynamic source–sink metapopulation structure, founder events and population bottlenecks. Evolution Biology. 2009;22(3):485–497. doi: 10.1111/j.1420-9101.2008.01675.x. - DOI - PubMed
-
- Beerli P. Migrate Documentation Version 3.2.1. Florida State University; Tallahassee, Florida: 2012. [8 September 2017].
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

