Novel putative drivers revealed by targeted exome sequencing of advanced solid tumors
- PMID: 29570743
- PMCID: PMC5865730
- DOI: 10.1371/journal.pone.0194790
Novel putative drivers revealed by targeted exome sequencing of advanced solid tumors
Abstract
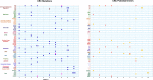
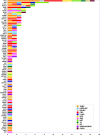
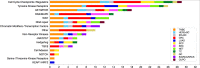
Next generation sequencing (NGS) is becoming increasingly integrated into oncological practice and clinical research. NGS methods have also provided evidence for clonal evolution of cancers during disease progression and treatment. The number of variants associated with response to specific therapeutic agents keeps increasing. However, the identification of novel driver mutations as opposed to passenger (phenotypically silent or clinically irrelevant) mutations remains a major challenge. We conducted targeted exome sequencing of advanced solid tumors from 44 pre-treated patients with solid tumors including breast, colorectal and lung carcinomas, neuroendocrine tumors, sarcomas and others. We catalogued established driver mutations and putative new drivers as predicted by two distinct algorithms. The established drivers we detected were consistent with published observations. However, we also detected a significant number of mutations with driver potential never described before in each tumor type we studied. These putative drivers belong to key cell fate regulatory networks, including potentially druggable pathways. Should our observations be confirmed, they would support the hypothesis that new driver mutations are selected by treatment in clinically aggressive tumors, and indicate a need for longitudinal genomic testing of solid tumors to inform second line cancer treatment.
Conflict of interest statement
Figures




References
-
- Moorcraft SY, Gonzalez D, Walker BA. Understanding next generation sequencing in oncology: A guide for oncologists. Critical reviews in oncology/hematology. 2015;96(3):463–74. Epub 2015/07/15. doi: 10.1016/j.critrevonc.2015.06.007 . - DOI - PubMed
-
- Carneiro BA, Costa R, Taxter T, Chandra S, Chae YK, Cristofanilli M, et al. Is Personalized Medicine Here? Oncology (Williston Park, NY). 2016;30(4):293–303, 7. Epub 2016/04/18. . - PubMed
-
- Nakagawa H, Wardell CP, Furuta M, Taniguchi H, Fujimoto A. Cancer whole-genome sequencing: present and future. Oncogene. 2015;34(49):5943–50. Epub 2015/03/31. doi: 10.1038/onc.2015.90 . - DOI - PubMed
-
- Ryu D, Joung JG, Kim NK, Kim KT, Park WY. Deciphering intratumor heterogeneity using cancer genome analysis. Human genetics. 2016;135(6):635–42. Epub 2016/04/30. doi: 10.1007/s00439-016-1670-x . - DOI - PubMed
-
- Shen T, Pajaro-Van de Stadt SH, Yeat NC, Lin JC. Clinical applications of next generation sequencing in cancer: from panels, to exomes, to genomes. Frontiers in genetics. 2015;6:215 Epub 2015/07/03. doi: 10.3389/fgene.2015.00215 ; PubMed Central PMCID: PMC4469892. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

