Genetic diversity and symbiotic effectiveness of Phaseolus vulgaris-nodulating rhizobia in Kenya
- PMID: 29571921
- PMCID: PMC6052332
- DOI: 10.1016/j.syapm.2018.02.001
Genetic diversity and symbiotic effectiveness of Phaseolus vulgaris-nodulating rhizobia in Kenya
Abstract
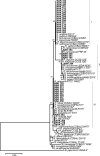
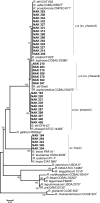
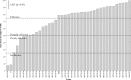
Phaseolus vulgaris (common bean) was introduced to Kenya several centuries ago but the rhizobia that nodulate it in the country remain poorly characterised. To address this gap in knowledge, 178 isolates recovered from the root nodules of P. vulgaris cultivated in Kenya were genotyped stepwise by the analysis of genomic DNA fingerprints, PCR-RFLP and 16S rRNA, atpD, recA and nodC gene sequences. Results indicated that P. vulgaris in Kenya is nodulated by at least six Rhizobium genospecies, with most of the isolates belonging to Rhizobium phaseoli and a possibly novel Rhizobium species. Infrequently, isolates belonged to Rhizobium paranaense, Rhizobium leucaenae, Rhizobium sophoriradicis and Rhizobium aegyptiacum. Despite considerable core-gene heterogeneity among the isolates, only four nodC gene alleles were observed indicating conservation within this gene. Testing of the capacity of the isolates to fix nitrogen (N2) in symbiosis with P. vulgaris revealed wide variations in effectiveness, with ten isolates comparable to Rhizobium tropici CIAT 899, a commercial inoculant strain for P. vulgaris. In addition to unveiling effective native rhizobial strains with potential as inoculants in Kenya, this study demonstrated that Kenyan soils harbour diverse P. vulgaris-nodulating rhizobia, some of which formed phylogenetic clusters distinct from known lineages. The native rhizobia differed by site, suggesting that field inoculation of P. vulgaris may need to be locally optimised.
Keywords: MLSA; Nodulation; Phaseolus vulgaris; Phylogeny; Rhizobium.
Copyright © 2018 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Figures

Similar articles
-
Phylogenetic diversity of rhizobial species and symbiovars nodulating Phaseolus vulgaris in Iran.FEMS Microbiol Lett. 2016 Mar;363(5):fnw024. doi: 10.1093/femsle/fnw024. Epub 2016 Jan 31. FEMS Microbiol Lett. 2016. PMID: 26832644
-
Genomic insight into the taxonomy of Rhizobium genospecies that nodulate Phaseolus vulgaris.Syst Appl Microbiol. 2018 Jul;41(4):300-310. doi: 10.1016/j.syapm.2018.03.001. Epub 2018 Mar 16. Syst Appl Microbiol. 2018. PMID: 29576402
-
Biodiversity and biogeography of rhizobia associated with common bean (Phaseolus vulgaris L.) in Shaanxi Province.Syst Appl Microbiol. 2016 May;39(3):211-219. doi: 10.1016/j.syapm.2016.02.001. Epub 2016 Feb 20. Syst Appl Microbiol. 2016. PMID: 26966063
-
The promiscuity of Phaseolus vulgaris L. (common bean) for nodulation with rhizobia: a review.World J Microbiol Biotechnol. 2020 Apr 20;36(5):63. doi: 10.1007/s11274-020-02839-w. World J Microbiol Biotechnol. 2020. PMID: 32314065 Review.
-
Genetic diversity of rhizobia associated with root nodules of white lupin (Lupinus albus L.) in Tunisian calcareous soils.Syst Appl Microbiol. 2019 Jul;42(4):448-456. doi: 10.1016/j.syapm.2019.04.002. Epub 2019 Apr 13. Syst Appl Microbiol. 2019. PMID: 31031015 Review.
Cited by
-
Phylogeography of the Bradyrhizobium spp. Associated With Peanut, Arachis hypogaea: Fellow Travelers or New Associations?Front Microbiol. 2019 Sep 4;10:2041. doi: 10.3389/fmicb.2019.02041. eCollection 2019. Front Microbiol. 2019. PMID: 31551977 Free PMC article.
-
Competition in the Phaseolus vulgaris-Rhizobium symbiosis and the role of resident soil rhizobia in determining the outcomes of inoculation.Plant Soil. 2023;487(1-2):61-77. doi: 10.1007/s11104-023-05903-0. Epub 2023 Feb 3. Plant Soil. 2023. PMID: 37333056 Free PMC article.
-
Microbial consortia inoculation of woody legume Erythrina brucei increases nodulation and shoot nitrogen and phosphorus under greenhouse conditions.Biotechnol Rep (Amst). 2022 Jan 31;33:e00707. doi: 10.1016/j.btre.2022.e00707. eCollection 2022 Mar. Biotechnol Rep (Amst). 2022. PMID: 35145889 Free PMC article.
-
Phylogenetic diversity of Rhizobium species recovered from nodules of common beans (Phaseolus vulgaris L.) in fields in Uganda: R. phaseoli, R. etli, and R. hidalgonense.FEMS Microbiol Ecol. 2024 Oct 25;100(11):fiae120. doi: 10.1093/femsec/fiae120. FEMS Microbiol Ecol. 2024. PMID: 39270668 Free PMC article.
-
Phylogeographic distribution of rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia.FEMS Microbiol Ecol. 2021 Apr 1;97(4):fiab046. doi: 10.1093/femsec/fiab046. FEMS Microbiol Ecol. 2021. PMID: 33724341 Free PMC article.
References
-
- Anyango B., Wilson K., Giller K. Competition in Kenyan soils between Rhizobium leguminosarum biovar phaseoli strain Kim5 and R. tropici strain CIAT 899 using the gusA marker gene. Plant Soil. 1998;204:69–78.
-
- Aserse A.A., Räsänen L.A., Assefa F., Hailemariam A., Lindström K. Phylogeny and genetic diversity of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia. Syst. Appl. Microbiol. 2012;35:120––131. - PubMed
-
- Asfaw A., Blair M.W., Almekinders C. Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theor. Appl. Genet. 2009;120:1––12. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

