Assessment of Passive Traps Combined with High-Throughput Sequencing To Study Airborne Fungal Communities
- PMID: 29572213
- PMCID: PMC5960964
- DOI: 10.1128/AEM.02637-17
Assessment of Passive Traps Combined with High-Throughput Sequencing To Study Airborne Fungal Communities
Abstract
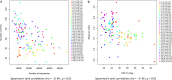
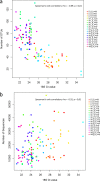
Techniques based on high-throughput sequencing (HTS) of environmental DNA have provided a new way of studying fungal diversity. However, these techniques suffer from a number of methodological biases which may appear at any of the steps involved in a metabarcoding study. Air is one of the most important environments where fungi can be found, because it is the primary medium of dispersal for many species. Looking ahead to future developments, it was decided to test 20 protocols, including different passive spore traps, spore recovery procedures, DNA extraction kits, and barcode loci. HTS was performed with the Illumina MiSeq platform targeting two subloci of the fungal internal transcribed spacer. Multivariate analysis and generalized linear models showed that the type of passive spore trap, the spore recovery procedure, and the barcode all impact the description of fungal communities in terms of richness and diversity when assessed by HTS metabarcoding. In contrast, DNA extraction kits did not significantly impact these results. Although passive traps may be used to describe airborne fungal communities, a study using specific real-time PCR and a mock community showed that these kinds of traps are affected by environmental conditions that may induce losses of biological material, impacting diversity and community composition results.IMPORTANCE The advent of high-throughput sequencing (HTS) methods, such as those offered by next-generation sequencing (NGS) techniques, has opened a new era in the study of fungal diversity in different environmental substrates. In this study, we show that an assessment of the diversity of airborne fungal communities can reliably be achieved by the use of simple and robust passive spore traps. However, a comparison of sample processing protocols showed that several methodological biases may impact the results of fungal diversity when assessed by metabarcoding. Our data suggest that identifying these biases is of paramount importance to enable a correct identification and relative quantification of community members.
Keywords: aerobiology; fungal dispersion; fungal diversity; metabarcoding.
Copyright © 2018 American Society for Microbiology.
Figures



References
-
- Desprez-Loustau M-L, Aguayo J, Dutech C, Hayden KJ, Husson C, Jakushkin B, Marçais B, Piou D, Robin C, Vacher C. 2016. An evolutionary ecology perspective to address forest pathology challenges of today and tomorrow. Ann Forest Sci 73:45–67. doi:10.1007/s13595-015-0487-4. - DOI
-
- Halme P, Heilmann-Clausen J, Rämä T, Kosonen T, Kunttu P. 2012. Monitoring fungal biodiversity–towards an integrated approach. Fungal Ecol 5:750–758. doi:10.1016/j.funeco.2012.05.005. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

