Comparative Proteomics of Dying and Surviving Cancer Cells Improves the Identification of Drug Targets and Sheds Light on Cell Life/Death Decisions
- PMID: 29572246
- PMCID: PMC5986252
- DOI: 10.1074/mcp.RA118.000610
Comparative Proteomics of Dying and Surviving Cancer Cells Improves the Identification of Drug Targets and Sheds Light on Cell Life/Death Decisions
Abstract
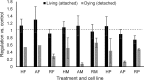
Chemotherapeutics cause the detachment and death of adherent cancer cells. When studying the proteome changes to determine the protein target and mechanism of action of anticancer drugs, the still-attached cells are normally used, whereas the detached cells are usually ignored. To test the hypothesis that proteomes of detached cells contain valuable information, we separately analyzed the proteomes of detached and attached HCT-116, A375, and RKO cells treated for 48 h with 5-fluorouracil, methotrexate and paclitaxel. Individually, the proteomic data on attached and detached cells had comparable performance in target and drug mechanism deconvolution, whereas the combined data significantly improved the target ranking for paclitaxel. Comparative analysis of attached versus detached proteomes provided further insight into cell life and death decision making. Six proteins consistently up- or downregulated in the detached versus attached cells regardless of the drug and cell type were discovered; their role in cell death/survival was tested by silencing them with siRNA. Knocking down USP11, CTTN, ACAA2, and EIF4H had anti-proliferative effects, affecting UHRF1 additionally sensitized the cells to the anticancer drugs, while knocking down RNF-40 increased cell survival against the treatments. Therefore, adding detached cells to the expression proteomics analysis of drug-treated cells can significantly increase the analytical value of the approach. The data have been deposited to the ProteomeXchange with identifier PXD007686.
Keywords: Cancer therapeutics; Cell death*; Cell survival; Drug resistance; Drug targets*; Label-free quantification; Mass Spectrometry; Mechanism of action; chemotherapeutic.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Lee J. A., Uhlik M. T., Moxham C. M., Tomandl D., and Sall D. J. (2012) Modern phenotypic drug discovery is a viable, neoclassic pharma strategy. J. Med. Chem. 55, 4527–4538 - PubMed
-
- Swinney D. C., and Anthony J. (2011) How were new medicines discovered? Nature reviews Drug Discovery 10, 507–519 - PubMed
-
- Eder J., Sedrani R., and Wiesmann C. (2014) The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug discovery 13, 577–587 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

