A genomics approach reveals insights into the importance of gene losses for mammalian adaptations
- PMID: 29572503
- PMCID: PMC5865188
- DOI: 10.1038/s41467-018-03667-1
A genomics approach reveals insights into the importance of gene losses for mammalian adaptations
Erratum in
-
Author Correction: A genomics approach reveals insights into the importance of gene losses for mammalian adaptations.Nat Commun. 2019 Dec 10;10(1):5707. doi: 10.1038/s41467-019-13828-5. Nat Commun. 2019. PMID: 31822665 Free PMC article.
Abstract
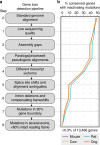
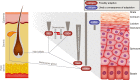
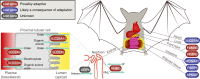
Identifying the genomic changes that underlie phenotypic adaptations is a key challenge in evolutionary biology and genomics. Loss of protein-coding genes is one type of genomic change with the potential to affect phenotypic evolution. Here, we develop a genomics approach to accurately detect gene losses and investigate their importance for adaptive evolution in mammals. We discover a number of gene losses that likely contributed to morphological, physiological, and metabolic adaptations in aquatic and flying mammals. These gene losses shed light on possible molecular and cellular mechanisms that underlie these adaptive phenotypes. In addition, we show that gene loss events that occur as a consequence of relaxed selection following adaptation provide novel insights into species' biology. Our results suggest that gene loss is an evolutionary mechanism for adaptation that may be more widespread than previously anticipated. Hence, investigating gene losses has great potential to reveal the genomic basis underlying macroevolutionary changes.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

