Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids
- PMID: 29572528
- PMCID: PMC5865108
- DOI: 10.1038/s41467-018-03469-5
Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids
Abstract
Cell-free protein synthesis has emerged as a powerful approach for expanding the range of genetically encoded chemistry into proteins. Unfortunately, efforts to site-specifically incorporate multiple non-canonical amino acids into proteins using crude extract-based cell-free systems have been limited by release factor 1 competition. Here we address this limitation by establishing a bacterial cell-free protein synthesis platform based on genomically recoded Escherichia coli lacking release factor 1. This platform was developed by exploiting multiplex genome engineering to enhance extract performance by functionally inactivating negative effectors. Our most productive cell extracts enabled synthesis of 1,780 ± 30 mg/L superfolder green fluorescent protein. Using an optimized platform, we demonstrated the ability to introduce 40 identical p-acetyl-L-phenylalanine residues site specifically into an elastin-like polypeptide with high accuracy of incorporation ( ≥ 98%) and yield (96 ± 3 mg/L). We expect this cell-free platform to facilitate fundamental understanding and enable manufacturing paradigms for proteins with new and diverse chemistries.
Conflict of interest statement
The authors declare no competing interests.
Figures


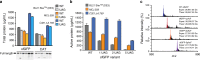
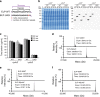
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

