Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study
- PMID: 29574065
- PMCID: PMC5968368
- DOI: 10.1016/S1473-3099(18)30073-2
Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study
Erratum in
-
Corrections.Lancet Infect Dis. 2018 May;18(5):492. doi: 10.1016/S1473-3099(18)30227-5. Epub 2018 Mar 27. Lancet Infect Dis. 2018. PMID: 29602752 Free PMC article. No abstract available.
Abstract
Background: In many countries, regular monitoring of the emergence of resistance to anti-tuberculosis drugs is hampered by the limitations of phenotypic testing for drug susceptibility. We therefore evaluated the use of genetic sequencing for surveillance of drug resistance in tuberculosis.
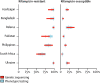
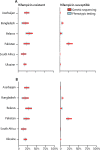
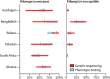
Methods: Population-level surveys were done in hospitals and clinics in seven countries (Azerbaijan, Bangladesh, Belarus, Pakistan, Philippines, South Africa, and Ukraine) to evaluate the use of genetic sequencing to estimate the resistance of Mycobacterium tuberculosis isolates to rifampicin, isoniazid, ofloxacin, moxifloxacin, pyrazinamide, kanamycin, amikacin, and capreomycin. For each drug, we assessed the accuracy of genetic sequencing by a comparison of the adjusted prevalence of resistance, measured by genetic sequencing, with the true prevalence of resistance, determined by phenotypic testing.
Findings: Isolates were taken from 7094 patients with tuberculosis who were enrolled in the study between November, 2009, and May, 2014. In all tuberculosis cases, the overall pooled sensitivity values for predicting resistance by genetic sequencing were 91% (95% CI 87-94) for rpoB (rifampicin resistance), 86% (74-93) for katG, inhA, and fabG promoter combined (isoniazid resistance), 54% (39-68) for pncA (pyrazinamide resistance), 85% (77-91) for gyrA and gyrB combined (ofloxacin resistance), and 88% (81-92) for gyrA and gyrB combined (moxifloxacin resistance). For nearly all drugs and in most settings, there was a large overlap in the estimated prevalence of drug resistance by genetic sequencing and the estimated prevalence by phenotypic testing.
Interpretation: Genetic sequencing can be a valuable tool for surveillance of drug resistance, providing new opportunities to monitor drug resistance in tuberculosis in resource-poor countries. Before its widespread adoption for surveillance purposes, there is a need to standardise DNA extraction methods, recording and reporting nomenclature, and data interpretation.
Funding: Bill & Melinda Gates Foundation, United States Agency for International Development, Global Alliance for Tuberculosis Drug Development.
© 2018 World Health Organization; licensee Elsevier. This is an Open Access article published under the CC BY 3.0 IGO license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In any use of this article, there should be no suggestion that WHO endorses any specific organisation, products or services. The use of the WHO logo is not permitted. This notice should be preserved along with the article's original URL.
Figures


Comment in
-
Population monitoring for drug-resistant tuberculosis: is genomics the answer?Lancet Infect Dis. 2018 Jun;18(6):592-594. doi: 10.1016/S1473-3099(18)30161-0. Epub 2018 Mar 21. Lancet Infect Dis. 2018. PMID: 29574064 No abstract available.
References
-
- WHO Antimicrobial resistance: global report on surveillance 2014. April, 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed Dec 29, 2017).
-
- WHO Global tuberculosis report 2017. 2017. http://www.who.int/tb/publications/global_report/en/ (accessed Dec 29, 2017).
-
- Zignol M, Dean AS, Falzon D. Twenty years of global surveillance of antituberculosis-drug resistance. N Engl J Med. 2016;375:1081–1089. - PubMed
-
- Uplekar M, Weil D, Lonnroth K. WHO's new end TB strategy. Lancet. 2015;385:1799–1801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

