Modifications generated by fast photochemical oxidation of proteins reflect the native conformations of proteins
- PMID: 29575296
- PMCID: PMC5980583
- DOI: 10.1002/pro.3408
Modifications generated by fast photochemical oxidation of proteins reflect the native conformations of proteins
Abstract
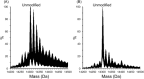
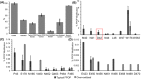
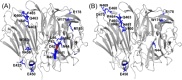
Hydroxyl radical footprinting (HRF) is a nonspecific protein footprinting method that has been increasingly used in recent years to analyze protein structure. The method oxidatively modifies solvent accessible sites in proteins, which changes upon alterations in the protein, such as ligand binding or a change in conformation. For HRF to provide accurate structural information, the method must probe the native structure of proteins. This requires careful experimental controls since an abundance of oxidative modifications can induce protein unfolding. Fast photochemical oxidation of proteins (FPOP) is a HRF method that generates hydroxyl radicals via photo-dissociation of hydrogen peroxide using an excimer laser. The addition of a radical scavenger to the FPOP reaction reduces the lifetime of the radical, limiting the levels of protein oxidation. A direct assay is needed to ensure FPOP is probing the native conformation of the protein. Here, we report using enzymatic activity as a direct assay to validate that FPOP is probing the native structure of proteins. By measuring the catalytic activity of lysozyme and invertase after FPOP modification, we demonstrate that FPOP does not induce protein unfolding.
Keywords: fast photochemical oxidation of protein (FPOP); hydroxyl radical footprinting (HRF); mass spectrometry; native protein structure; oxidative modifications; protein footprinting.
© 2018 The Protein Society.
Figures



Similar articles
-
Fast photochemical oxidation of proteins (FPOP): A powerful mass spectrometry-based structural proteomics tool.J Biol Chem. 2019 Aug 9;294(32):11969-11979. doi: 10.1074/jbc.REV119.006218. Epub 2019 Jul 1. J Biol Chem. 2019. PMID: 31262727 Free PMC article. Review.
-
Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization.Acc Chem Res. 2018 Mar 20;51(3):736-744. doi: 10.1021/acs.accounts.7b00593. Epub 2018 Feb 16. Acc Chem Res. 2018. PMID: 29450991 Free PMC article. Review.
-
Characterizing Cellular Proteins with In-cell Fast Photochemical Oxidation of Proteins.J Vis Exp. 2020 Mar 11;(157). doi: 10.3791/60911. J Vis Exp. 2020. PMID: 32225159
-
Fast Photochemical Oxidation of Proteins Coupled with Mass Spectrometry.Protein Pept Lett. 2019;26(1):27-34. doi: 10.2174/0929866526666181128124554. Protein Pept Lett. 2019. PMID: 30484399 Free PMC article. Review.
-
In Vivo Hydroxyl Radical Protein Footprinting for the Study of Protein Interactions in Caenorhabditis elegans.J Vis Exp. 2020 Apr 1;(158):10.3791/60910. doi: 10.3791/60910. J Vis Exp. 2020. PMID: 32310230 Free PMC article.
Cited by
-
Characterizing Thermal Transitions of IgG with Mass Spectrometry.J Am Soc Mass Spectrom. 2019 Nov;30(11):2438-2445. doi: 10.1007/s13361-019-02292-6. Epub 2019 Jul 30. J Am Soc Mass Spectrom. 2019. PMID: 31363989 Free PMC article.
-
Benefits of Ion Mobility Separation and Parallel Accumulation-Serial Fragmentation Technology on timsTOF Pro for the Needs of Fast Photochemical Oxidation of Protein Analysis.ACS Omega. 2021 Apr 8;6(15):10352-10361. doi: 10.1021/acsomega.1c00732. eCollection 2021 Apr 20. ACS Omega. 2021. PMID: 34056188 Free PMC article.
-
Fast photochemical oxidation of proteins (FPOP): A powerful mass spectrometry-based structural proteomics tool.J Biol Chem. 2019 Aug 9;294(32):11969-11979. doi: 10.1074/jbc.REV119.006218. Epub 2019 Jul 1. J Biol Chem. 2019. PMID: 31262727 Free PMC article. Review.
-
In-Cell Labeling and Mass Spectrometry for Systems-Level Structural Biology.Chem Rev. 2022 Apr 27;122(8):7647-7689. doi: 10.1021/acs.chemrev.1c00223. Epub 2021 Jul 7. Chem Rev. 2022. PMID: 34232610 Free PMC article. Review.
-
Evolution of Structural Biology through the Lens of Mass Spectrometry.Anal Chem. 2019 Jan 2;91(1):142-155. doi: 10.1021/acs.analchem.8b05014. Epub 2018 Dec 6. Anal Chem. 2019. PMID: 30457831 Free PMC article. Review. No abstract available.
References
-
- Kiselar JG, Janmey PA, Almo SC, Chance MR (2003) Structural analysis of gelsolin using synchrotron protein footprinting. Mol Cell Proteomics 2:1120–1132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

